序言和数据集
1 %matplotlib notebook 2 import numpy as np 3 import pandas as pd 4 import seaborn as sn 5 import matplotlib.pyplot as plt 6 7 from sklearn.model_selection import train_test_split 8 from sklearn.datasets import make_classification, make_blobs 9 from matplotlib.colors import ListedColormap 10 from sklearn.datasets import load_breast_cancer 11 from adspy_shared_utilities import load_crime_dataset 12 13 14 cmap_bold = ListedColormap(['#FFFF00', '#00FF00', '#0000FF','#000000']) 15 16 # fruits dataset 17 fruits = pd.read_table('fruit_data_with_colors.txt') 18 19 feature_names_fruits = ['height', 'width', 'mass', 'color_score'] 20 X_fruits = fruits[feature_names_fruits] 21 y_fruits = fruits['fruit_label'] 22 target_names_fruits = ['apple', 'mandarin', 'orange', 'lemon'] 23 24 X_fruits_2d = fruits[['height', 'width']] 25 y_fruits_2d = fruits['fruit_label'] 26 27 # synthetic dataset for simple regression 28 from sklearn.datasets import make_regression 29 plt.figure() 30 plt.title('Sample regression problem with one input variable') 31 X_R1, y_R1 = make_regression(n_samples = 100, n_features=1, 32 n_informative=1, bias = 150.0, 33 noise = 30, random_state=0) 34 plt.scatter(X_R1, y_R1, marker= 'o', s=50) 35 plt.show() 36 37 # synthetic dataset for more complex regression 38 from sklearn.datasets import make_friedman1 39 plt.figure() 40 plt.title('Complex regression problem with one input variable') 41 X_F1, y_F1 = make_friedman1(n_samples = 100, n_features = 7, 42 random_state=0) 43 44 plt.scatter(X_F1[:, 2], y_F1, marker= 'o', s=50) 45 plt.show() 46 47 # synthetic dataset for classification (binary) 48 plt.figure() 49 plt.title('Sample binary classification problem with two informative features') 50 X_C2, y_C2 = make_classification(n_samples = 100, n_features=2, 51 n_redundant=0, n_informative=2, 52 n_clusters_per_class=1, flip_y = 0.1, 53 class_sep = 0.5, random_state=0) 54 plt.scatter(X_C2[:, 0], X_C2[:, 1], marker= 'o', 55 c=y_C2, s=50, cmap=cmap_bold) 56 plt.show() 57 58 # more difficult synthetic dataset for classification (binary) 59 # with classes that are not linearly separable 60 X_D2, y_D2 = make_blobs(n_samples = 100, n_features = 2, 61 centers = 8, cluster_std = 1.3, 62 random_state = 4) 63 y_D2 = y_D2 % 2 64 plt.figure() 65 plt.title('Sample binary classification problem with non-linearly separable classes') 66 plt.scatter(X_D2[:,0], X_D2[:,1], c=y_D2, 67 marker= 'o', s=50, cmap=cmap_bold) 68 plt.show() 69 70 # Breast cancer dataset for classification 71 cancer = load_breast_cancer() 72 (X_cancer, y_cancer) = load_breast_cancer(return_X_y = True) 73 74 # Communities and Crime dataset 75 (X_crime, y_crime) = load_crime_dataset()



贝叶斯分类器
1 from sklearn.naive_bayes import GaussianNB 2 from adspy_shared_utilities import plot_class_regions_for_classifier 3 4 X_train, X_test, y_train, y_test = train_test_split(X_C2, y_C2, random_state=0) 5 6 nbclf = GaussianNB().fit(X_train, y_train) 7 plot_class_regions_for_classifier(nbclf, X_train, y_train, X_test, y_test, 8 'Gaussian Naive Bayes classifier: Dataset 1')

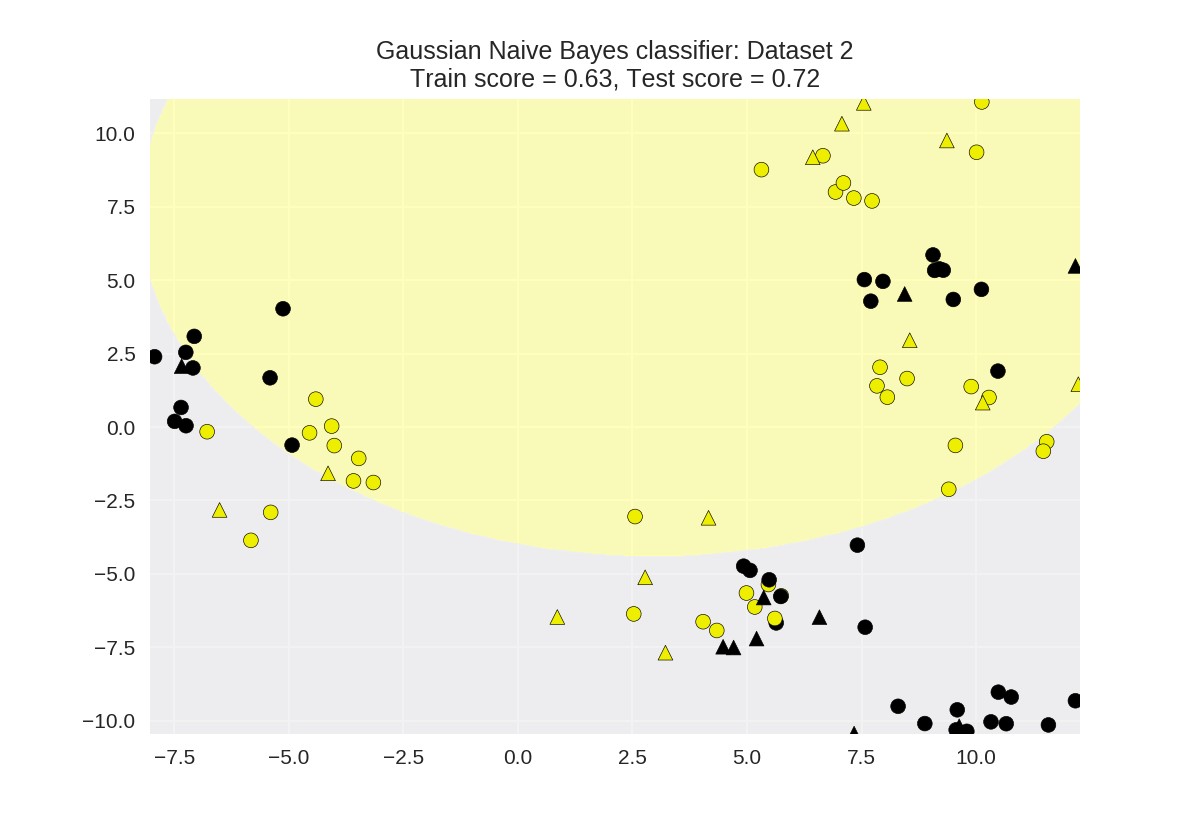
1 X_train, X_test, y_train, y_test = train_test_split(X_D2, y_D2, 2 random_state=0) 3 4 nbclf = GaussianNB().fit(X_train, y_train) 5 plot_class_regions_for_classifier(nbclf, X_train, y_train, X_test, y_test, 6 'Gaussian Naive Bayes classifier: Dataset 2')
Application to a real-world dataset
1 X_train, X_test, y_train, y_test = train_test_split(X_cancer, y_cancer, random_state = 0) 2 3 nbclf = GaussianNB().fit(X_train, y_train) 4 print('Breast cancer dataset') 5 print('Accuracy of GaussianNB classifier on training set: {:.2f}' 6 .format(nbclf.score(X_train, y_train))) 7 print('Accuracy of GaussianNB classifier on test set: {:.2f}' 8 .format(nbclf.score(X_test, y_test)))
Breast cancer dataset Accuracy of GaussianNB classifier on training set: 0.95 Accuracy of GaussianNB classifier on test set: 0.94
决策树集成
随机森林
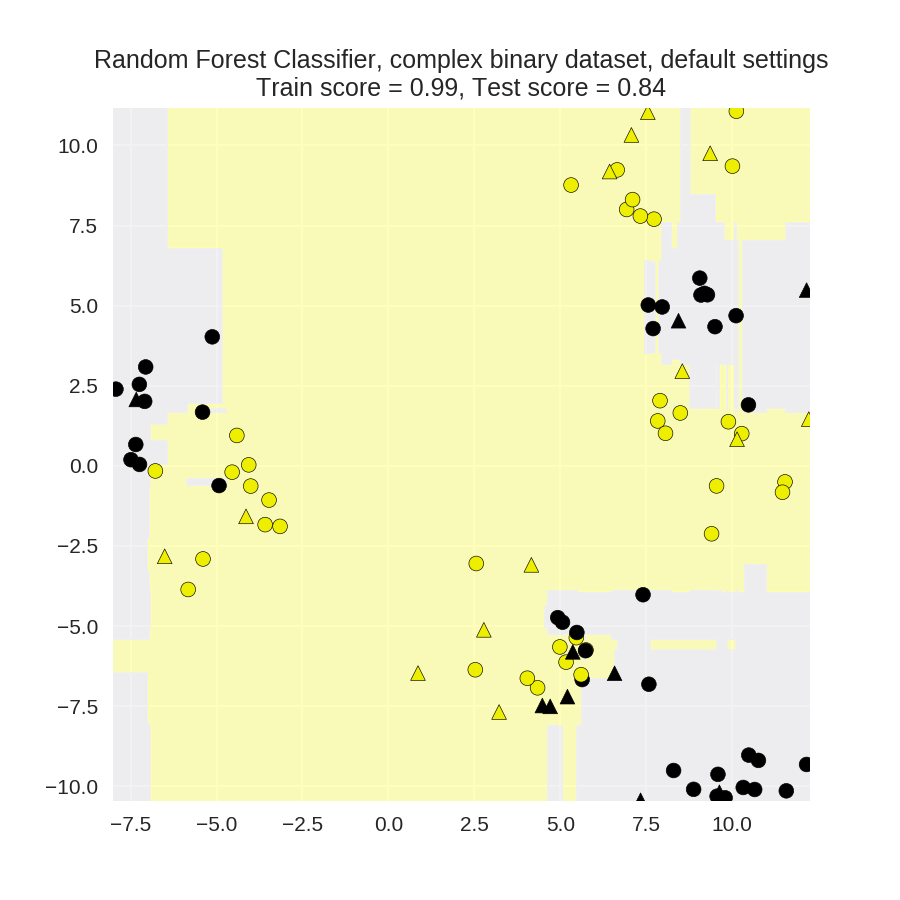
1 from sklearn.ensemble import RandomForestClassifier 2 from sklearn.model_selection import train_test_split 3 from adspy_shared_utilities import plot_class_regions_for_classifier_subplot 4 5 X_train, X_test, y_train, y_test = train_test_split(X_D2, y_D2, 6 random_state = 0) 7 fig, subaxes = plt.subplots(1, 1, figsize=(6, 6)) 8 9 clf = RandomForestClassifier().fit(X_train, y_train) 10 title = 'Random Forest Classifier, complex binary dataset, default settings' 11 plot_class_regions_for_classifier_subplot(clf, X_train, y_train, X_test, 12 y_test, title, subaxes) 13 14 plt.show()
Random forest: Fruit dataset
1 from sklearn.ensemble import RandomForestClassifier 2 from sklearn.model_selection import train_test_split 3 from adspy_shared_utilities import plot_class_regions_for_classifier_subplot 4 5 X_train, X_test, y_train, y_test = train_test_split(X_fruits.as_matrix(), 6 y_fruits.as_matrix(), 7 random_state = 0) 8 fig, subaxes = plt.subplots(6, 1, figsize=(6, 32)) 9 10 title = 'Random Forest, fruits dataset, default settings' 11 pair_list = [[0,1], [0,2], [0,3], [1,2], [1,3], [2,3]] 12 13 for pair, axis in zip(pair_list, subaxes): 14 X = X_train[:, pair] 15 y = y_train 16 17 clf = RandomForestClassifier().fit(X, y) 18 plot_class_regions_for_classifier_subplot(clf, X, y, None, 19 None, title, axis, 20 target_names_fruits) 21 22 axis.set_xlabel(feature_names_fruits[pair[0]]) 23 axis.set_ylabel(feature_names_fruits[pair[1]]) 24 25 plt.tight_layout() 26 plt.show() 27 28 clf = RandomForestClassifier(n_estimators = 10, 29 random_state=0).fit(X_train, y_train) 30 31 print('Random Forest, Fruit dataset, default settings') 32 print('Accuracy of RF classifier on training set: {:.2f}' 33 .format(clf.score(X_train, y_train))) 34 print('Accuracy of RF classifier on test set: {:.2f}' 35 .format(clf.score(X_test, y_test)))

Random Forest, Fruit dataset, default settings Accuracy of RF classifier on training set: 1.00 Accuracy of RF classifier on test set: 0.80
Random Forests on a real-world dataset
1 from sklearn.ensemble import RandomForestClassifier 2 3 X_train, X_test, y_train, y_test = train_test_split(X_cancer, y_cancer, random_state = 0) 4 5 clf = RandomForestClassifier(max_features = 8, random_state = 0) 6 clf.fit(X_train, y_train) 7 8 print('Breast cancer dataset') 9 print('Accuracy of RF classifier on training set: {:.2f}' 10 .format(clf.score(X_train, y_train))) 11 print('Accuracy of RF classifier on test set: {:.2f}' 12 .format(clf.score(X_test, y_test)))
Breast cancer dataset Accuracy of RF classifier on training set: 1.00 Accuracy of RF classifier on test set: 0.99
Gradient-boosted decision trees
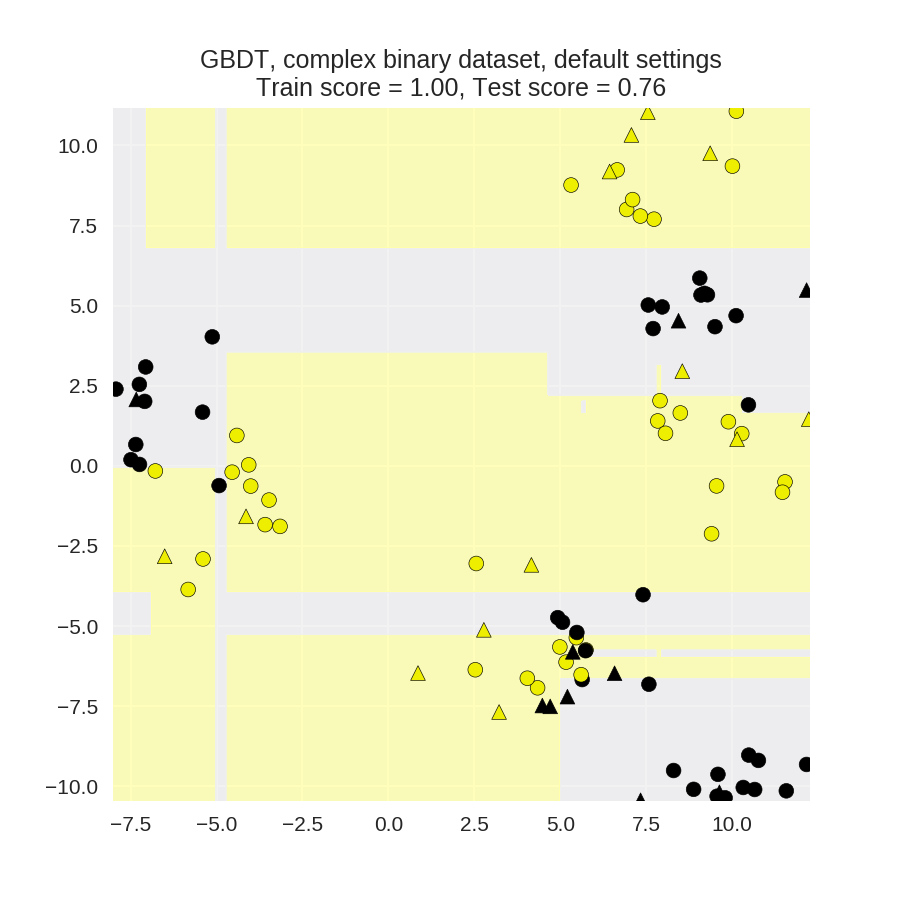
1 from sklearn.ensemble import GradientBoostingClassifier 2 from sklearn.model_selection import train_test_split 3 from adspy_shared_utilities import plot_class_regions_for_classifier_subplot 4 5 X_train, X_test, y_train, y_test = train_test_split(X_D2, y_D2, random_state = 0) 6 fig, subaxes = plt.subplots(1, 1, figsize=(6, 6)) 7 8 clf = GradientBoostingClassifier().fit(X_train, y_train) 9 title = 'GBDT, complex binary dataset, default settings' 10 plot_class_regions_for_classifier_subplot(clf, X_train, y_train, X_test, 11 y_test, title, subaxes) 12 13 plt.show()
Gradient boosted decision trees on the fruit dataset
1 X_train, X_test, y_train, y_test = train_test_split(X_fruits.as_matrix(), 2 y_fruits.as_matrix(), 3 random_state = 0) 4 fig, subaxes = plt.subplots(6, 1, figsize=(6, 32)) 5 6 pair_list = [[0,1], [0,2], [0,3], [1,2], [1,3], [2,3]] 7 8 for pair, axis in zip(pair_list, subaxes): 9 X = X_train[:, pair] 10 y = y_train 11 12 clf = GradientBoostingClassifier().fit(X, y) 13 plot_class_regions_for_classifier_subplot(clf, X, y, None, 14 None, title, axis, 15 target_names_fruits) 16 17 axis.set_xlabel(feature_names_fruits[pair[0]]) 18 axis.set_ylabel(feature_names_fruits[pair[1]]) 19 20 plt.tight_layout() 21 plt.show() 22 clf = GradientBoostingClassifier().fit(X_train, y_train) 23 24 print('GBDT, Fruit dataset, default settings') 25 print('Accuracy of GBDT classifier on training set: {:.2f}' 26 .format(clf.score(X_train, y_train))) 27 print('Accuracy of GBDT classifier on test set: {:.2f}' 28 .format(clf.score(X_test, y_test)))

GBDT, Fruit dataset, default settings Accuracy of GBDT classifier on training set: 1.00 Accuracy of GBDT classifier on test set: 0.80
Gradient-boosted decision trees on a real-world dataset
1 from sklearn.ensemble import GradientBoostingClassifier 2 3 X_train, X_test, y_train, y_test = train_test_split(X_cancer, y_cancer, random_state = 0) 4 5 clf = GradientBoostingClassifier(random_state = 0) 6 clf.fit(X_train, y_train) 7 8 print('Breast cancer dataset (learning_rate=0.1, max_depth=3)') 9 print('Accuracy of GBDT classifier on training set: {:.2f}' 10 .format(clf.score(X_train, y_train))) 11 print('Accuracy of GBDT classifier on test set: {:.2f}\n' 12 .format(clf.score(X_test, y_test))) 13 14 clf = GradientBoostingClassifier(learning_rate = 0.01, max_depth = 2, random_state = 0) 15 clf.fit(X_train, y_train) 16 17 print('Breast cancer dataset (learning_rate=0.01, max_depth=2)') 18 print('Accuracy of GBDT classifier on training set: {:.2f}' 19 .format(clf.score(X_train, y_train))) 20 print('Accuracy of GBDT classifier on test set: {:.2f}' 21 .format(clf.score(X_test, y_test)))
神经网络
几种常见激励函数(能够进行非线性你决策)
1 xrange = np.linspace(-2, 2, 200) 2 3 plt.figure(figsize=(7,6)) 4 5 plt.plot(xrange, np.maximum(xrange, 0), label = 'relu') 6 plt.plot(xrange, np.tanh(xrange), label = 'tanh') 7 plt.plot(xrange, 1 / (1 + np.exp(-xrange)), label = 'logistic') 8 plt.legend() 9 plt.title('Neural network activation functions') 10 plt.xlabel('Input value (x)') 11 plt.ylabel('Activation function output') 12 13 plt.show()

神经网络:分类
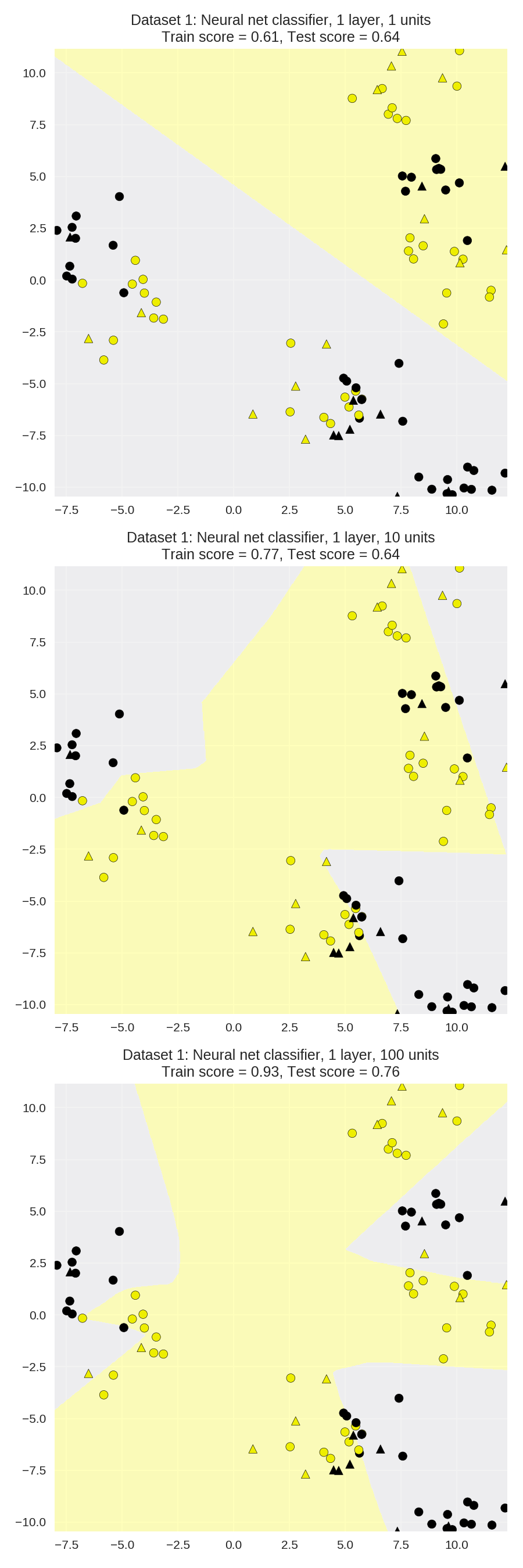
Synthetic dataset 1: single hidden layer(单一隐含层)
1 from sklearn.neural_network import MLPClassifier 2 from adspy_shared_utilities import plot_class_regions_for_classifier_subplot 3 4 X_train, X_test, y_train, y_test = train_test_split(X_D2, y_D2, random_state=0) 5 6 fig, subaxes = plt.subplots(3, 1, figsize=(6,18)) 7 8 for units, axis in zip([1, 10, 100], subaxes): 9 nnclf = MLPClassifier(hidden_layer_sizes = [units], solver='lbfgs', 10 random_state = 0).fit(X_train, y_train) 11 12 title = 'Dataset 1: Neural net classifier, 1 layer, {} units'.format(units) 13 14 plot_class_regions_for_classifier_subplot(nnclf, X_train, y_train, 15 X_test, y_test, title, axis) 16 plt.tight_layout()
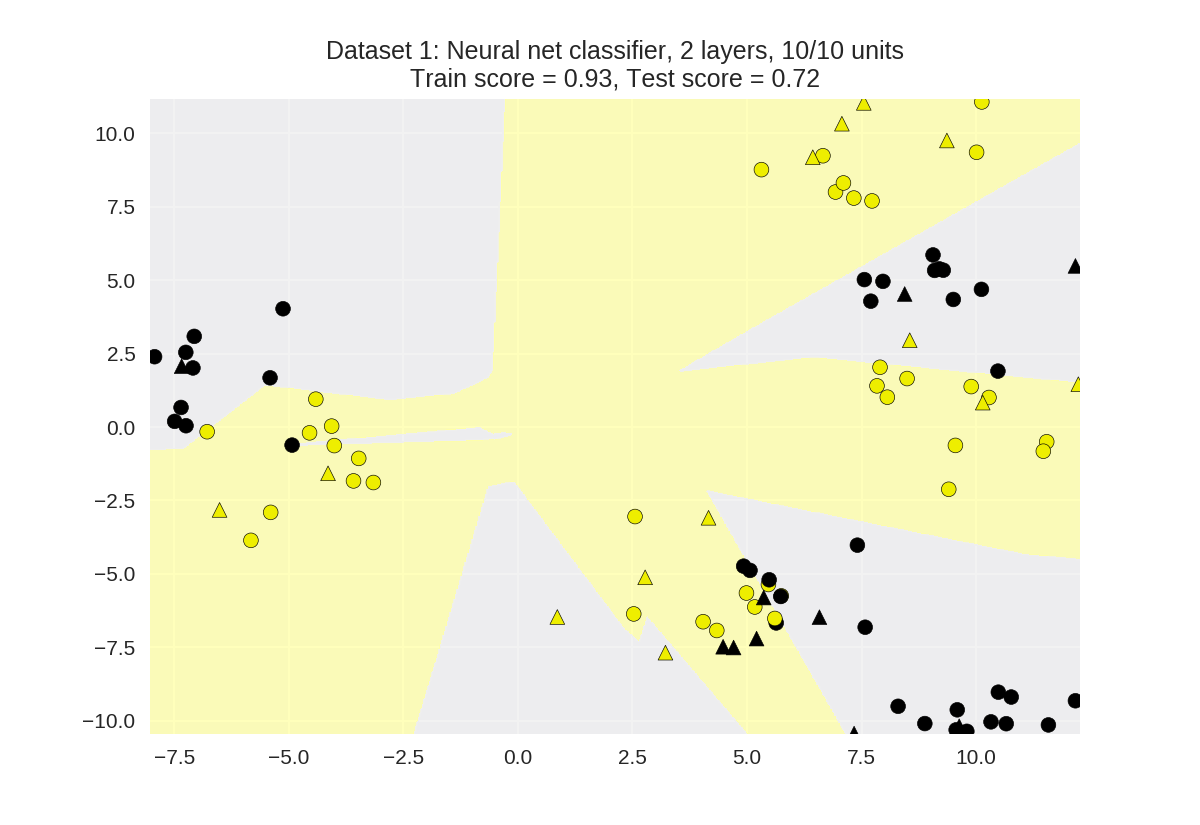
双隐含层
1 from adspy_shared_utilities import plot_class_regions_for_classifier 2 3 X_train, X_test, y_train, y_test = train_test_split(X_D2, y_D2, random_state=0) 4 5 nnclf = MLPClassifier(hidden_layer_sizes = [10, 10], solver='lbfgs', 6 random_state = 0).fit(X_train, y_train) 7 8 plot_class_regions_for_classifier(nnclf, X_train, y_train, X_test, y_test, 9 'Dataset 1: Neural net classifier, 2 layers, 10/10 units')
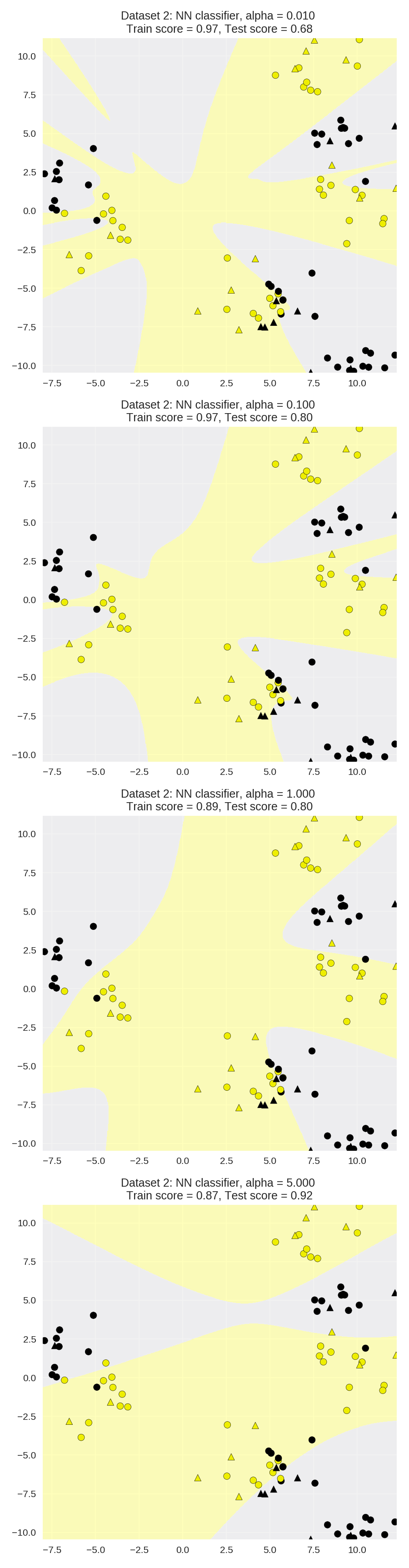
Regularization parameter: alpha
1 X_train, X_test, y_train, y_test = train_test_split(X_D2, y_D2, random_state=0) 2 3 fig, subaxes = plt.subplots(4, 1, figsize=(6, 23)) 4 5 for this_alpha, axis in zip([0.01, 0.1, 1.0, 5.0], subaxes): 6 nnclf = MLPClassifier(solver='lbfgs', activation = 'tanh', 7 alpha = this_alpha, 8 hidden_layer_sizes = [100, 100], 9 random_state = 0).fit(X_train, y_train) 10 11 title = 'Dataset 2: NN classifier, alpha = {:.3f} '.format(this_alpha) 12 13 plot_class_regions_for_classifier_subplot(nnclf, X_train, y_train, 14 X_test, y_test, title, axis) 15 plt.tight_layout() 16
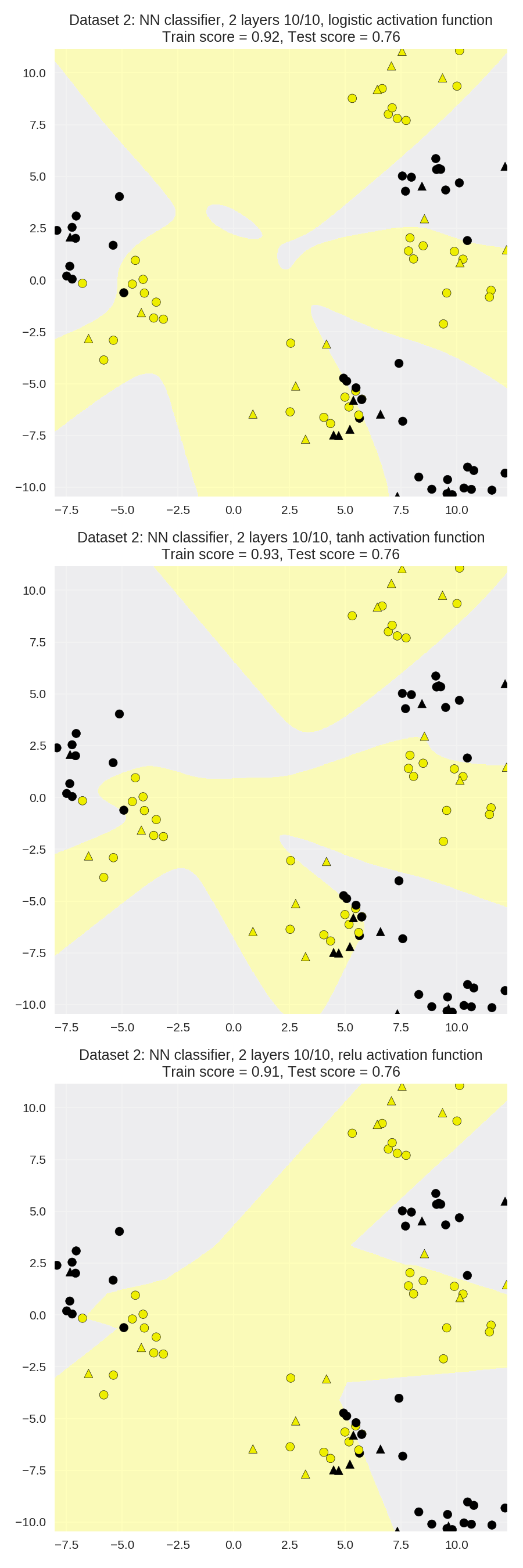
The effect of different choices of activation function
1 X_train, X_test, y_train, y_test = train_test_split(X_D2, y_D2, random_state=0) 2 3 fig, subaxes = plt.subplots(3, 1, figsize=(6,18)) 4 5 for this_activation, axis in zip(['logistic', 'tanh', 'relu'], subaxes): 6 nnclf = MLPClassifier(solver='lbfgs', activation = this_activation, 7 alpha = 0.1, hidden_layer_sizes = [10, 10], 8 random_state = 0).fit(X_train, y_train) 9 10 title = 'Dataset 2: NN classifier, 2 layers 10/10, {} \ 11 activation function'.format(this_activation) 12 13 plot_class_regions_for_classifier_subplot(nnclf, X_train, y_train, 14 X_test, y_test, title, axis) 15 plt.tight_layout()
神经网络:回归
1 from sklearn.neural_network import MLPRegressor 2 3 fig, subaxes = plt.subplots(2, 3, figsize=(11,8), dpi=70) 4 5 X_predict_input = np.linspace(-3, 3, 50).reshape(-1,1) 6 7 X_train, X_test, y_train, y_test = train_test_split(X_R1[0::5], y_R1[0::5], random_state = 0) 8 9 for thisaxisrow, thisactivation in zip(subaxes, ['tanh', 'relu']): 10 for thisalpha, thisaxis in zip([0.0001, 1.0, 100], thisaxisrow): 11 mlpreg = MLPRegressor(hidden_layer_sizes = [100,100], 12 activation = thisactivation, 13 alpha = thisalpha, 14 solver = 'lbfgs').fit(X_train, y_train) 15 y_predict_output = mlpreg.predict(X_predict_input) 16 thisaxis.set_xlim([-2.5, 0.75]) 17 thisaxis.plot(X_predict_input, y_predict_output, 18 '^', markersize = 10) 19 thisaxis.plot(X_train, y_train, 'o') 20 thisaxis.set_xlabel('Input feature') 21 thisaxis.set_ylabel('Target value') 22 thisaxis.set_title('MLP regression\nalpha={}, activation={})' 23 .format(thisalpha, thisactivation)) 24 plt.tight_layout()

Application to real-world dataset for classification
1 from sklearn.neural_network import MLPClassifier 2 from sklearn.preprocessing import MinMaxScaler 3 4 5 scaler = MinMaxScaler() 6 7 X_train, X_test, y_train, y_test = train_test_split(X_cancer, y_cancer, random_state = 0) 8 X_train_scaled = scaler.fit_transform(X_train) 9 X_test_scaled = scaler.transform(X_test) 10 11 clf = MLPClassifier(hidden_layer_sizes = [100, 100], alpha = 5.0, 12 random_state = 0, solver='lbfgs').fit(X_train_scaled, y_train) 13 14 print('Breast cancer dataset') 15 print('Accuracy of NN classifier on training set: {:.2f}' 16 .format(clf.score(X_train_scaled, y_train))) 17 print('Accuracy of NN classifier on test set: {:.2f}' 18 .format(clf.score(X_test_scaled, y_test)))
Breast cancer dataset Accuracy of NN classifier on training set: 0.98 Accuracy of NN classifier on test set: 0.97



