



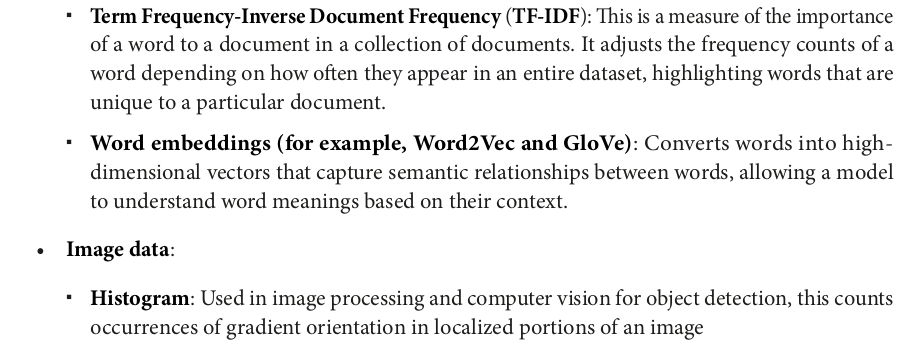

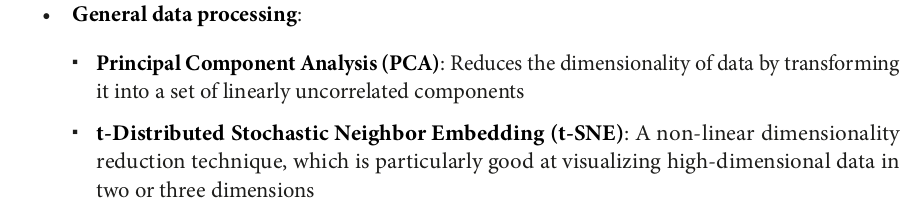


from pyspark.sql import SparkSession from pyspark.ml.feature import HashingTF, IDF, Tokenizer spark = SparkSession.builder.appName("TF-IDF Example").getOrCreate() data = [ (0, "This is the first document"), (1, "This document is the second document"), (2, "And this is the third one"), (3, "Is this the first document?"), (4, "The last document is the fifth one") ] df = spark.createDataFrame(data, ["id", "text"]) df.show(truncate=False)
+---+------------------------------------+ |id |text | +---+------------------------------------+ |0 |This is the first document | |1 |This document is the second document| |2 |And this is the third one | |3 |Is this the first document? | |4 |The last document is the fifth one | +---+------------------------------------+
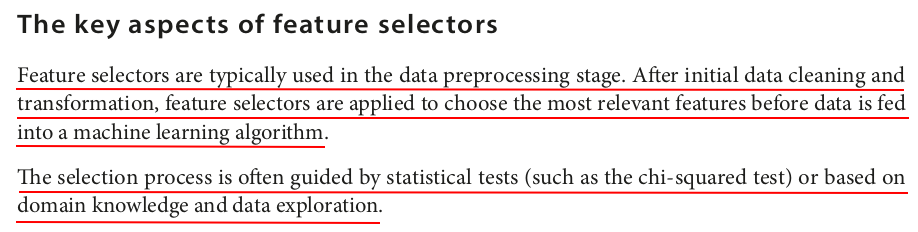
We tokenize the text using Tokenizer:
tokenizer = Tokenizer(inputCol="text", outputCol="words") words_df = tokenizer.transform(df) words_df.show(truncate=False)
+---+------------------------------------+-------------------------------------------+ |id |text |words | +---+------------------------------------+-------------------------------------------+ |0 |This is the first document |[this, is, the, first, document] | |1 |This document is the second document|[this, document, is, the, second, document]| |2 |And this is the third one |[and, this, is, the, third, one] | |3 |Is this the first document? |[is, this, the, first, document?] | |4 |The last document is the fifth one |[the, last, document, is, the, fifth, one] | +---+------------------------------------+-------------------------------------------+
We apply HashingTF to convert words into feature vectors:
hashingTF = HashingTF(inputCol="words", outputCol="rawFeatures", numFeatures=15) featurized_df = hashingTF.transform(words_df) featurized_df.show(truncate=False)
+---+------------------------------------+-------------------------------------------+--------------------------------------------+ |id |text |words |rawFeatures | +---+------------------------------------+-------------------------------------------+--------------------------------------------+ |0 |This is the first document |[this, is, the, first, document] |(15,[2,3,4,10,13],[1.0,1.0,1.0,1.0,1.0]) | |1 |This document is the second document|[this, document, is, the, second, document]|(15,[2,3,4,6,10],[1.0,1.0,1.0,1.0,2.0]) | |2 |And this is the third one |[and, this, is, the, third, one] |(15,[0,1,2,3,4,6],[1.0,1.0,1.0,1.0,1.0,1.0])| |3 |Is this the first document? |[is, this, the, first, document?] |(15,[1,2,3,4,13],[1.0,1.0,1.0,1.0,1.0]) | |4 |The last document is the fifth one |[the, last, document, is, the, fifth, one] |(15,[0,2,4,9,10],[1.0,3.0,1.0,1.0,1.0]) | +---+------------------------------------+-------------------------------------------+--------------------------------------------+
We compute the IDF using IDF:
idf = IDF(inputCol="rawFeatures", outputCol="features") idf_model = idf.fit(featurized_df) tfidf_df = idf_model.transform(featurized_df) tfidf_df.show(truncate=False)
+---+------------------------------------+-------------------------------------------+--------------------------------------------+--------------------------------------------------------------------------------------------------------+ |id |text |words |rawFeatures |features | +---+------------------------------------+-------------------------------------------+--------------------------------------------+--------------------------------------------------------------------------------------------------------+ |0 |This is the first document |[this, is, the, first, document] |(15,[2,3,4,10,13],[1.0,1.0,1.0,1.0,1.0]) |(15,[2,3,4,10,13],[0.0,0.1823215567939546,0.0,0.4054651081081644,0.6931471805599453]) | |1 |This document is the second document|[this, document, is, the, second, document]|(15,[2,3,4,6,10],[1.0,1.0,1.0,1.0,2.0]) |(15,[2,3,4,6,10],[0.0,0.1823215567939546,0.0,0.6931471805599453,0.8109302162163288]) | |2 |And this is the third one |[and, this, is, the, third, one] |(15,[0,1,2,3,4,6],[1.0,1.0,1.0,1.0,1.0,1.0])|(15,[0,1,2,3,4,6],[0.6931471805599453,0.6931471805599453,0.0,0.1823215567939546,0.0,0.6931471805599453])| |3 |Is this the first document? |[is, this, the, first, document?] |(15,[1,2,3,4,13],[1.0,1.0,1.0,1.0,1.0]) |(15,[1,2,3,4,13],[0.6931471805599453,0.0,0.1823215567939546,0.0,0.6931471805599453]) | |4 |The last document is the fifth one |[the, last, document, is, the, fifth, one] |(15,[0,2,4,9,10],[1.0,3.0,1.0,1.0,1.0]) |(15,[0,2,4,9,10],[0.6931471805599453,0.0,0.0,1.0986122886681098,0.4054651081081644]) | +---+------------------------------------+-------------------------------------------+--------------------------------------------+--------------------------------------------------------------------------------------------------------+
Finally, we select the relevant columns, including the TF-IDF features.
result_df = tfidf_df.select("id", "text", "features") result_df.show(truncate=False,vertical=False)
+---+------------------------------------+--------------------------------------------------------------------------------------------------------+ |id |text |features | +---+------------------------------------+--------------------------------------------------------------------------------------------------------+ |0 |This is the first document |(15,[2,3,4,10,13],[0.0,0.1823215567939546,0.0,0.4054651081081644,0.6931471805599453]) | |1 |This document is the second document|(15,[2,3,4,6,10],[0.0,0.1823215567939546,0.0,0.6931471805599453,0.8109302162163288]) | |2 |And this is the third one |(15,[0,1,2,3,4,6],[0.6931471805599453,0.6931471805599453,0.0,0.1823215567939546,0.0,0.6931471805599453])| |3 |Is this the first document? |(15,[1,2,3,4,13],[0.6931471805599453,0.0,0.1823215567939546,0.0,0.6931471805599453]) | |4 |The last document is the fifth one |(15,[0,2,4,9,10],[0.6931471805599453,0.0,0.0,1.0986122886681098,0.4054651081081644]) | +---+------------------------------------+--------------------------------------------------------------------------------------------------------+

from pyspark.sql import SparkSession from pyspark.ml.feature import Word2Vec # Create a Spark session spark = SparkSession.builder.appName("Word2Vec Example").getOrCreate() # Sample data (documents with tokenized words) data = [ (0, ["apple", "banana", "orange", "grape"]), (1, ["apple", "banana", "cherry", "pear"]), (2, ["banana", "cherry", "grape", "kiwi"]), (3, ["apple", "pear", "kiwi", "orange"]), (4, ["cherry", "grape", "kiwi", "orange"]), ] # Create a DataFrame from the sample data df = spark.createDataFrame(data, ["id", "words"]) df.show(truncate=False)
+---+------------------------------+ |id |words | +---+------------------------------+ |0 |[apple, banana, orange, grape]| |1 |[apple, banana, cherry, pear] | |2 |[banana, cherry, grape, kiwi] | |3 |[apple, pear, kiwi, orange] | |4 |[cherry, grape, kiwi, orange] | +---+------------------------------+
The vectorSize parameter was set to 3, meaning that each word would be represented by a vector of three dimensions.
word2vec = Word2Vec(vectorSize=3, minCount=0,inputCol="words", outputCol="features")

model = word2vec.fit(df) result = model.transform(df) result.select("id", "features").show(truncate=False)
+---+------------------------------------------------------------------+ |id |features | +---+------------------------------------------------------------------+ |0 |[-0.03166230674833059,-0.014463053084909916,-0.039787358487956226]| |1 |[0.002557281404733658,-0.09344123024493456,-0.125989543274045] | |2 |[0.00833282619714737,0.01878553256392479,-0.08785744372289628] | |3 |[-0.06233153026551008,-0.06857131328433752,-0.09306064527481794] | |4 |[-0.013947931118309498,-0.012707299552857876,-0.06643945060204715]| +---+------------------------------------------------------------------+
We can interpret that each row (document) now has a vector of Word2Vec features. These embeddings can be useful for various NLP tasks, such as finding synonyms, clustering related words, or even training downstream models.
from pyspark.sql import SparkSession from pyspark.ml.feature import CountVectorizer spark = SparkSession.builder.appName("CountVectorizer Example").getOrCreate() data = [ (0, ["apple", "banana", "orange", "grape"]), (1, ["apple", "banana", "cherry", "pear"]), (2, ["banana", "cherry", "grape", "kiwi"]), (3, ["apple", "pear", "kiwi", "orange"]), (4, ["cherry", "grape", "kiwi", "orange"]), ] df = spark.createDataFrame(data, ["id", "words"]) df.show(truncate=False)
+---+------------------------------+ |id |words | +---+------------------------------+ |0 |[apple, banana, orange, grape]| |1 |[apple, banana, cherry, pear] | |2 |[banana, cherry, grape, kiwi] | |3 |[apple, pear, kiwi, orange] | |4 |[cherry, grape, kiwi, orange] | +---+------------------------------+

Each row (document) now has a vector of word counts:
cv = CountVectorizer(inputCol="words",outputCol="features", vocabSize=7) model = cv.fit(df) result = model.transform(df) result.select("id", "features").show(truncate=False)
+---+-------------------------------+ |id |features | +---+-------------------------------+ |0 |(7,[0,3,4,5],[1.0,1.0,1.0,1.0])| |1 |(7,[2,3,4,6],[1.0,1.0,1.0,1.0])| |2 |(7,[1,2,3,5],[1.0,1.0,1.0,1.0])| |3 |(7,[0,1,4,6],[1.0,1.0,1.0,1.0])| |4 |(7,[0,1,2,5],[1.0,1.0,1.0,1.0])| +---+-------------------------------+


from pyspark.sql import SparkSession from pyspark.ml.feature import FeatureHasher spark = SparkSession.builder.appName("FeatureHasher Example").getOrCreate() data = [ (0, "apple", "banana", "orange"), (1, "apple", "banana", "cherry"), (2, "banana", "cherry", "grape"), (3, "apple", "pear", "kiwi"), (4, "cherry", "grape", "kiwi"), ] df = spark.createDataFrame(data, ["id", "feature1", "feature2", "feature3"]) df.show(truncate=False)
+---+--------+--------+--------+ |id |feature1|feature2|feature3| +---+--------+--------+--------+ |0 |apple |banana |orange | |1 |apple |banana |cherry | |2 |banana |cherry |grape | |3 |apple |pear |kiwi | |4 |cherry |grape |kiwi | +---+--------+--------+--------+

hasher = FeatureHasher(inputCols=["feature1", "feature2", "feature3"],outputCol="features", numFeatures=10) result = hasher.transform(df) result.select("id","features").show(truncate=False)
+---+--------------------------+ |id |features | +---+--------------------------+ |0 |(10,[3,7,9],[1.0,1.0,1.0])| |1 |(10,[3,6,7],[1.0,1.0,1.0])| |2 |(10,[0,4,7],[1.0,1.0,1.0])| |3 |(10,[0,7,8],[1.0,1.0,1.0])| |4 |(10,[0,2,6],[1.0,1.0,1.0])| +---+--------------------------+


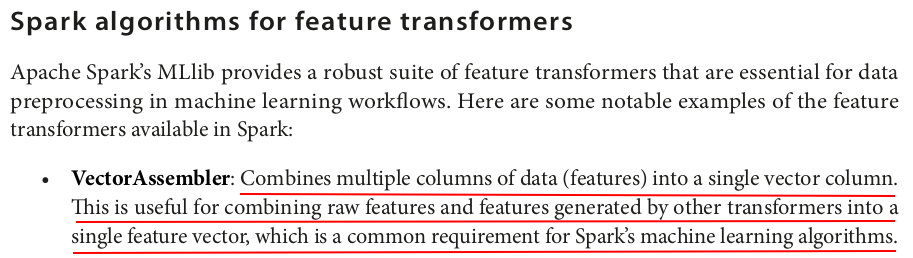


from pyspark.sql import SparkSession from pyspark.ml.feature import VectorAssembler spark = SparkSession.builder.appName("VectorAssembler Example").getOrCreate() data = [ (1, 2, 3, 4), (5, 6, 7, 8), (9, 10, 11, 12) ] columns = ["feature1", "feature2", "feature3", "feature4"] df = spark.createDataFrame(data, columns) df.show()
+--------+--------+--------+--------+ |feature1|feature2|feature3|feature4| +--------+--------+--------+--------+ | 1| 2| 3| 4| | 5| 6| 7| 8| | 9| 10| 11| 12| +--------+--------+--------+--------+
VectorAssembler is a feature transformer in PySpark that combines a given list of columns into a single vector column:
assembler = VectorAssembler(inputCols=columns, outputCol="features") output_df = assembler.transform(df) output_df.show()
+--------+--------+--------+--------+--------------------+ |feature1|feature2|feature3|feature4| features| +--------+--------+--------+--------+--------------------+ | 1| 2| 3| 4| [1.0,2.0,3.0,4.0]| | 5| 6| 7| 8| [5.0,6.0,7.0,8.0]| | 9| 10| 11| 12|[9.0,10.0,11.0,12.0]| +--------+--------+--------+--------+--------------------+

from pyspark.ml.feature import StringIndexer data = [("X", 100), ("X", 200), ("Y", 300), ("Y", 200), ("Y", 300), ("C", 400), ("Z", 100), ("Z", 100)] columns = ["Categories", "Value"] df = spark.createDataFrame(data, columns) df.show()
+----------+-----+ |Categories|Value| +----------+-----+ | X| 100| | X| 200| | Y| 300| | Y| 200| | Y| 300| | C| 400| | Z| 100| | Z| 100| +----------+-----+
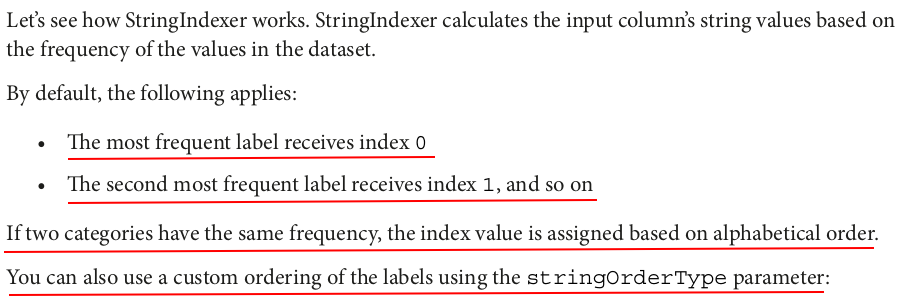
indexer = StringIndexer(inputCol="Categories",outputCol="Categories_Indexed") indexerModel = indexer.fit(df) indexed_df = indexerModel.transform(df) indexed_df.show()
+----------+-----+------------------+ |Categories|Value|Categories_Indexed| +----------+-----+------------------+ | X| 100| 1.0| | X| 200| 1.0| | Y| 300| 0.0| | Y| 200| 0.0| | Y| 300| 0.0| | C| 400| 3.0| | Z| 100| 2.0| | Z| 100| 2.0| +----------+-----+------------------+

from pyspark.ml.feature import OneHotEncoder data = [(0.0, 1.0), (1.0, 0.0), (2.0, 1.0)] columns = ["input1", "input2"] df = spark.createDataFrame(data, columns) df.show()
+------+------+ |input1|input2| +------+------+ | 0.0| 1.0| | 1.0| 0.0| | 2.0| 1.0| +------+------+
The following code snippet processes the input columns by applying the OneHotEncoder model. It then transforms the input columns.
encoder = OneHotEncoder( inputCols=["input1", "input2"], outputCols=["output1", "output2"] ) encoded_df = encoder.fit(df) encoded_df = encoded_df.transform(df) encoded_df.show(truncate=False)
+------+------+-------------+-------------+ |input1|input2|output1 |output2 | +------+------+-------------+-------------+ |0.0 |1.0 |(2,[0],[1.0])|(1,[],[]) | |1.0 |0.0 |(2,[1],[1.0])|(1,[0],[1.0])| |2.0 |1.0 |(2,[],[]) |(1,[],[]) | +------+------+-------------+-------------+

from pyspark.ml.feature import Tokenizer, RegexTokenizer data = [("Th+is is a sam+ple sent+ence.",)] columns = ["text"] df = spark.createDataFrame(data, columns) df.show(truncate=False)
+-----------------------------+ |text | +-----------------------------+ |Th+is is a sam+ple sent+ence.| +-----------------------------+

tokenizer = Tokenizer(inputCol="text", outputCol="tokens") tokenized_df = tokenizer.transform(df) regex_tokenizer = RegexTokenizer(inputCol="text",outputCol="regex_tokens", pattern="\\+") regex_tokenized_df = regex_tokenizer.transform(df) tokenized_df.show(truncate=False) regex_tokenized_df.show(truncate=False)
+-----------------------------+-----------------------------------+ |text |tokens | +-----------------------------+-----------------------------------+ |Th+is is a sam+ple sent+ence.|[th+is, is, a, sam+ple, sent+ence.]| +-----------------------------+-----------------------------------+ +-----------------------------+----------------------------------+ |text |regex_tokens | +-----------------------------+----------------------------------+ |Th+is is a sam+ple sent+ence.|[th, is is a sam, ple sent, ence.]| +-----------------------------+----------------------------------+

from pyspark.ml.feature import StopWordsRemover, Tokenizer data = [("This is the first sentence.",), ("And here's another sentence.",), ("A third sentence for the DataFrame.",)] columns = ["text"] df = spark.createDataFrame(data, columns) tokenizer = Tokenizer(inputCol="text", outputCol="words") df = tokenizer.transform(df) df.show(truncate=False)
+-----------------------------------+------------------------------------------+ |text |words | +-----------------------------------+------------------------------------------+ |This is the first sentence. |[this, is, the, first, sentence.] | |And here's another sentence. |[and, here's, another, sentence.] | |A third sentence for the DataFrame.|[a, third, sentence, for, the, dataframe.]| +-----------------------------------+------------------------------------------+
StopWordsRemover removes common stop words (“This,” “is,” “the,” “And,” “another”, “A”, and “for”) from the input sentence.
stopwords_remover = StopWordsRemover(inputCol="words", outputCol="filtered_words") filtered_df = stopwords_remover.transform(df) filtered_df.show(truncate=False)
+-----------------------------------+------------------------------------------+-----------------------------+ |text |words |filtered_words | +-----------------------------------+------------------------------------------+-----------------------------+ |This is the first sentence. |[this, is, the, first, sentence.] |[first, sentence.] | |And here's another sentence. |[and, here's, another, sentence.] |[another, sentence.] | |A third sentence for the DataFrame.|[a, third, sentence, for, the, dataframe.]|[third, sentence, dataframe.]| +-----------------------------------+------------------------------------------+-----------------------------+

from pyspark.ml.feature import Bucketizer from pyspark.sql.functions import col data = [ (0, 1.5), (1, 2.5), (2, 3.5), (3, 4.5), (4, 5.5) ] columns = ["id", "value"] df = spark.createDataFrame(data, columns) df.show()
+---+-----+ | id|value| +---+-----+ | 0| 1.5| | 1| 2.5| | 2| 3.5| | 3| 4.5| | 4| 5.5| +---+-----+
The following snippet applies the bucketizer function to transform data:
splits = [0.0, 2.0, 4.0, float("inf")] bucketizer = Bucketizer(splits=splits, inputCol="value",outputCol="bucket") bucketized_df = bucketizer.transform(df) bucketized_df.show()
+---+-----+------+ | id|value|bucket| +---+-----+------+ | 0| 1.5| 0.0| | 1| 2.5| 1.0| | 2| 3.5| 1.0| | 3| 4.5| 2.0| | 4| 5.5| 2.0| +---+-----+------+
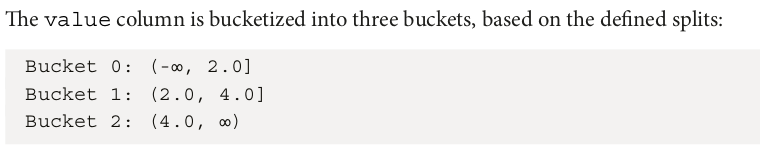

from pyspark.ml.feature import StandardScaler from pyspark.ml.linalg import Vectors data = [ (0, Vectors.dense([1.0, 0.1, -1.0]),), (1, Vectors.dense([2.0, 1.1, 1.0]),), (2, Vectors.dense([3.0, 10.1, 3.0]),) ] columns = ["id", "features"] df = spark.createDataFrame(data, columns) df.show()
+---+--------------+ | id| features| +---+--------------+ | 0|[1.0,0.1,-1.0]| | 1| [2.0,1.1,1.0]| | 2|[3.0,10.1,3.0]| +---+--------------+
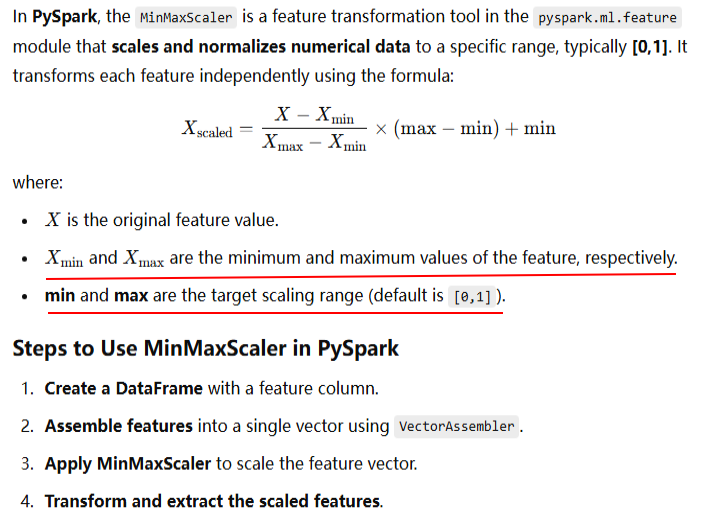
The following code snippet transforms the input columns into scaled features by applying the StandardScaler function:
scaler = StandardScaler(inputCol="features", outputCol="scaled_features",withStd=True, withMean=True) scaler_model = scaler.fit(df) scaled_df = scaler_model.transform(df) scaled_df.show(truncate=False)
+---+--------------+-------------------------------+ |id |features |scaled_features | +---+--------------+-------------------------------+ |0 |[1.0,0.1,-1.0]|[-1.0,-0.6657502859356826,-1.0]| |1 |[2.0,1.1,1.0] |[0.0,-0.4841820261350419,0.0] | |2 |[3.0,10.1,3.0]|[1.0,1.1499323120707245,1.0] | +---+--------------+-------------------------------+

from pyspark.ml.feature import MinMaxScaler from pyspark.ml.linalg import Vectors data = [ (0, Vectors.dense([1.0, 0.1, -1.0]),), (1, Vectors.dense([2.0, 1.1, 1.0]),), (2, Vectors.dense([3.0, 10.1, 3.0]),) ] columns = ["id", "features"] df = spark.createDataFrame(data, columns) df.show()
+---+--------------+ | id| features| +---+--------------+ | 0|[1.0,0.1,-1.0]| | 1| [2.0,1.1,1.0]| | 2|[3.0,10.1,3.0]| +---+--------------+
The following code snippet transforms the input column by applying the MinMaxScaler function:
scaler = MinMaxScaler(inputCol="features", outputCol="scaled_features") scaler_model = scaler.fit(df) scaled_df = scaler_model.transform(df) scaled_df.show(truncate=False)
+---+--------------+---------------+
|id |features |scaled_features|
+---+--------------+---------------+
|0 |[1.0,0.1,-1.0]|(3,[],[]) |
|1 |[2.0,1.1,1.0] |[0.5,0.1,0.5] |
|2 |[3.0,10.1,3.0]|[1.0,1.0,1.0] |
+---+--------------+---------------+

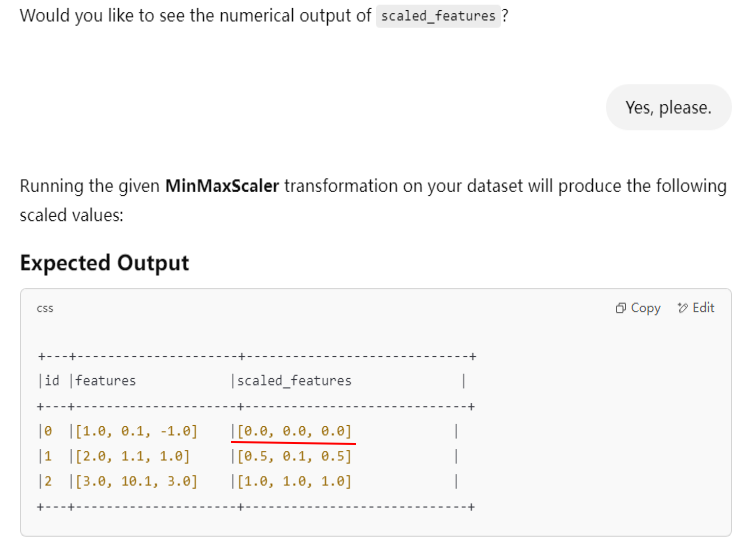
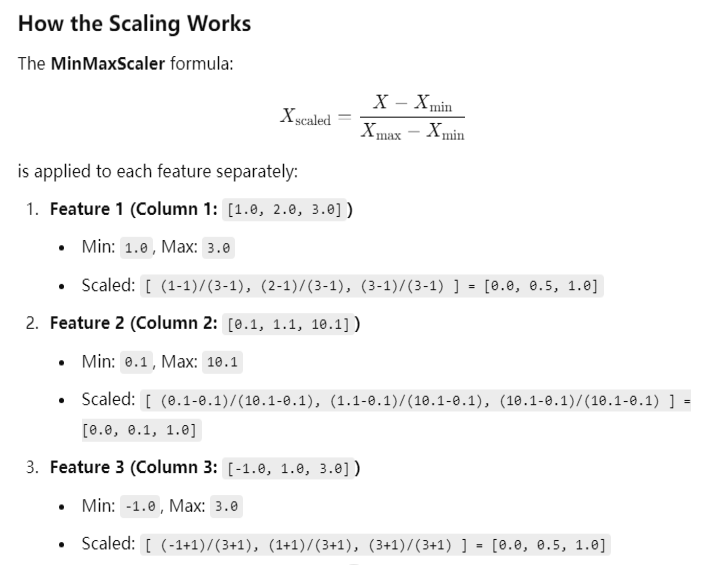
Tried another PySpark version 3.4.4, still got the wrong result.
The result is correct if divide the features column into three feature columns and apply MinMaxScaler for each:
from pyspark.ml.feature import MinMaxScaler from pyspark.ml.linalg import Vectors data = [ (0, Vectors.dense([1.0]),), (1, Vectors.dense([2.0]),), (2, Vectors.dense([3.0]),) ] columns = ["id", "features"] df = spark.createDataFrame(data, columns) scaler = MinMaxScaler(inputCol="features", outputCol="scaled_features") scaler_model = scaler.fit(df) scaled_df = scaler_model.transform(df) scaled_df.show(truncate=False)
+---+--------+---------------+ |id |features|scaled_features| +---+--------+---------------+ |0 |[1.0] |[0.0] | |1 |[2.0] |[0.5] | |2 |[3.0] |[1.0] | +---+--------+---------------+
data = [ (0, Vectors.dense([0.1]),), (1, Vectors.dense([1.1]),), (2, Vectors.dense([10.1]),) ] columns = ["id", "features"] df = spark.createDataFrame(data, columns) scaler = MinMaxScaler(inputCol="features", outputCol="scaled_features") scaler_model = scaler.fit(df) scaled_df = scaler_model.transform(df) scaled_df.show(truncate=False)
+---+--------+---------------+ |id |features|scaled_features| +---+--------+---------------+ |0 |[0.1] |[0.0] | |1 |[1.1] |[0.1] | |2 |[10.1] |[1.0] | +---+--------+---------------+
data = [ (0, Vectors.dense([-1.0]),), (1, Vectors.dense([1.0]),), (2, Vectors.dense([3.0]),) ] columns = ["id", "features"] df = spark.createDataFrame(data, columns) scaler = MinMaxScaler(inputCol="features", outputCol="scaled_features") scaler_model = scaler.fit(df) scaled_df = scaler_model.transform(df) scaled_df.show(truncate=False)
+---+--------+---------------+ |id |features|scaled_features| +---+--------+---------------+ |0 |[-1.0] |[0.0] | |1 |[1.0] |[0.5] | |2 |[3.0] |[1.0] | +---+--------+---------------+
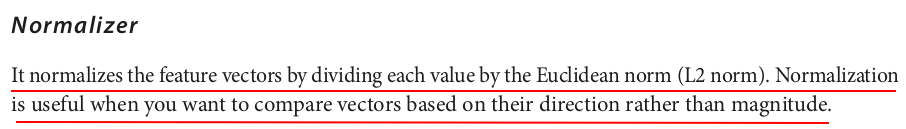
from pyspark.ml.feature import Normalizer from pyspark.ml.linalg import Vectors data = [ (0, Vectors.dense([1.0, 0.1, -1.0]),), (1, Vectors.dense([2.0, 1.1, 1.0]),), (2, Vectors.dense([3.0, 10.1, 3.0]),) ] columns = ["id", "features"] df = spark.createDataFrame(data, columns) normalizer = Normalizer(inputCol="features", outputCol="normalized_features", p=2.0) normalized_df = normalizer.transform(df) normalized_df.show(truncate=False)
+---+--------------+------------------------------------------------------------+ |id |features |normalized_features | +---+--------------+------------------------------------------------------------+ |0 |[1.0,0.1,-1.0]|[0.7053456158585983,0.07053456158585983,-0.7053456158585983]| |1 |[2.0,1.1,1.0] |[0.8025723539051279,0.4414147946478204,0.40128617695256397] | |2 |[3.0,10.1,3.0]|[0.27384986857909926,0.9219612242163009,0.27384986857909926]| +---+--------------+------------------------------------------------------------+
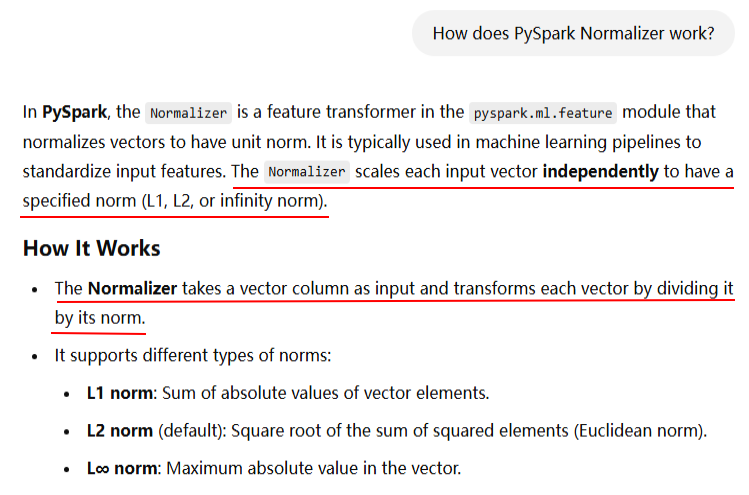
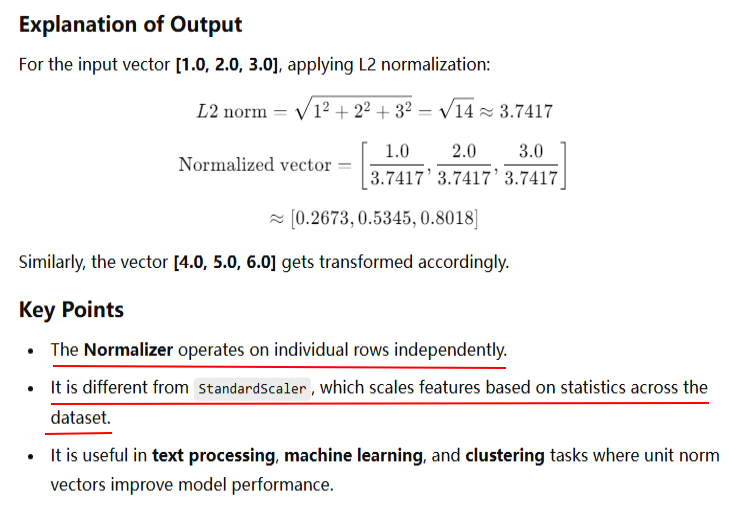
from pyspark.sql import SparkSession from pyspark.ml.feature import Normalizer from pyspark.ml.linalg import Vectors # Initialize Spark Session spark = SparkSession.builder.appName("NormalizerExample").getOrCreate() # Sample Data data = [(0, Vectors.dense([1.0, 2.0, 3.0])), (1, Vectors.dense([4.0, 5.0, 6.0]))] df = spark.createDataFrame(data, ["id", "features"]) # Initialize Normalizer normalizer = Normalizer(inputCol="features", outputCol="normalized_features", p=2.0) # L2 norm # Transform Data normalized_df = normalizer.transform(df) # Show Results normalized_df.show(truncate=False) # Stop Spark Session spark.stop()
data = [ (0, Vectors.dense([1.0]),), (1, Vectors.dense([2.0]),), (2, Vectors.dense([3.0]),) ] columns = ["id", "features"] df = spark.createDataFrame(data, columns) normalizer = Normalizer(inputCol="features", outputCol="normalized_features", p=2.0) normalized_df = normalizer.transform(df) normalized_df.show(truncate=False)
+---+--------+-------------------+ |id |features|normalized_features| +---+--------+-------------------+ |0 |[1.0] |[1.0] | |1 |[2.0] |[1.0] | |2 |[3.0] |[1.0] | +---+--------+-------------------+

from pyspark.ml.feature import PCA from pyspark.ml.linalg import Vectors data = [ (0, Vectors.dense([1.0, 0.1, -1.0]),), (1, Vectors.dense([2.0, 1.1, 1.0]),), (2, Vectors.dense([3.0, 10.1, 3.0]),) ] columns = ["id", "features"] df = spark.createDataFrame(data, columns) pca = PCA(k=2, inputCol="features", outputCol="pca_features") pca_model = pca.fit(df) pca_df = pca_model.transform(df) pca_df.show(truncate=False)
+---+--------------+------------------------------------------+ |id |features |pca_features | +---+--------------+------------------------------------------+ |0 |[1.0,0.1,-1.0]|[0.06466700238304013,-0.45367188451874657]| |1 |[2.0,1.1,1.0] |[-1.6616789696362084,1.284065030233573] | |2 |[3.0,10.1,3.0]|[-10.870750062210382,0.19181523649833365] | +---+--------------+------------------------------------------+
The PCA model reduces the dimensionality of the features column to two principal components.
data = [ (0, Vectors.dense([1.0, 0.1, -1.0]),) ] columns = ["id", "features"] df = spark.createDataFrame(data, columns) pca = PCA(k=2, inputCol="features", outputCol="pca_features") pca_model = pca.fit(df) pca_df = pca_model.transform(df) pca_df.show(truncate=False)
--------------------------------------------------------------------------- IllegalArgumentException Traceback (most recent call last) Cell In[10], line 11 8 df = spark.createDataFrame(data, columns) 10 pca = PCA(k=2, inputCol="features", outputCol="pca_features") ---> 11 pca_model = pca.fit(df) 12 pca_df = pca_model.transform(df) 13 pca_df.show(truncate=False) File /zdata/anaconda3/lib/python3.12/site-packages/pyspark/ml/base.py:205, in Estimator.fit(self, dataset, params) 203 return self.copy(params)._fit(dataset) 204 else: --> 205 return self._fit(dataset) 206 else: 207 raise TypeError( 208 "Params must be either a param map or a list/tuple of param maps, " 209 "but got %s." % type(params) 210 ) File /zdata/anaconda3/lib/python3.12/site-packages/pyspark/ml/wrapper.py:381, in JavaEstimator._fit(self, dataset) 380 def _fit(self, dataset: DataFrame) -> JM: --> 381 java_model = self._fit_java(dataset) 382 model = self._create_model(java_model) 383 return self._copyValues(model) File /zdata/anaconda3/lib/python3.12/site-packages/pyspark/ml/wrapper.py:378, in JavaEstimator._fit_java(self, dataset) 375 assert self._java_obj is not None 377 self._transfer_params_to_java() --> 378 return self._java_obj.fit(dataset._jdf) File /zdata/anaconda3/lib/python3.12/site-packages/py4j/java_gateway.py:1322, in JavaMember.__call__(self, *args) 1316 command = proto.CALL_COMMAND_NAME +\ 1317 self.command_header +\ 1318 args_command +\ 1319 proto.END_COMMAND_PART 1321 answer = self.gateway_client.send_command(command) -> 1322 return_value = get_return_value( 1323 answer, self.gateway_client, self.target_id, self.name) 1325 for temp_arg in temp_args: 1326 if hasattr(temp_arg, "_detach"): File /zdata/anaconda3/lib/python3.12/site-packages/pyspark/errors/exceptions/captured.py:185, in capture_sql_exception.<locals>.deco(*a, **kw) 181 converted = convert_exception(e.java_exception) 182 if not isinstance(converted, UnknownException): 183 # Hide where the exception came from that shows a non-Pythonic 184 # JVM exception message. --> 185 raise converted from None 186 else: 187 raise IllegalArgumentException: requirement failed: RowMatrix.computeCovariance called on matrix with only 1 rows. Cannot compute the covariance of a RowMatrix with <= 1 row.
data = [ (0, Vectors.dense([1.0]),), (1, Vectors.dense([2.0]),), (2, Vectors.dense([3.0]),) ] columns = ["id", "features"] df = spark.createDataFrame(data, columns) pca = PCA(k=2, inputCol="features", outputCol="pca_features") pca_model = pca.fit(df) pca_df = pca_model.transform(df) pca_df.show(truncate=False)
--------------------------------------------------------------------------- IllegalArgumentException Traceback (most recent call last) Cell In[11], line 10 7 df = spark.createDataFrame(data, columns) 9 pca = PCA(k=2, inputCol="features", outputCol="pca_features") ---> 10 pca_model = pca.fit(df) 11 pca_df = pca_model.transform(df) 12 pca_df.show(truncate=False) File /zdata/anaconda3/lib/python3.12/site-packages/pyspark/ml/base.py:205, in Estimator.fit(self, dataset, params) 203 return self.copy(params)._fit(dataset) 204 else: --> 205 return self._fit(dataset) 206 else: 207 raise TypeError( 208 "Params must be either a param map or a list/tuple of param maps, " 209 "but got %s." % type(params) 210 ) File /zdata/anaconda3/lib/python3.12/site-packages/pyspark/ml/wrapper.py:381, in JavaEstimator._fit(self, dataset) 380 def _fit(self, dataset: DataFrame) -> JM: --> 381 java_model = self._fit_java(dataset) 382 model = self._create_model(java_model) 383 return self._copyValues(model) File /zdata/anaconda3/lib/python3.12/site-packages/pyspark/ml/wrapper.py:378, in JavaEstimator._fit_java(self, dataset) 375 assert self._java_obj is not None 377 self._transfer_params_to_java() --> 378 return self._java_obj.fit(dataset._jdf) File /zdata/anaconda3/lib/python3.12/site-packages/py4j/java_gateway.py:1322, in JavaMember.__call__(self, *args) 1316 command = proto.CALL_COMMAND_NAME +\ 1317 self.command_header +\ 1318 args_command +\ 1319 proto.END_COMMAND_PART 1321 answer = self.gateway_client.send_command(command) -> 1322 return_value = get_return_value( 1323 answer, self.gateway_client, self.target_id, self.name) 1325 for temp_arg in temp_args: 1326 if hasattr(temp_arg, "_detach"): File /zdata/anaconda3/lib/python3.12/site-packages/pyspark/errors/exceptions/captured.py:185, in capture_sql_exception.<locals>.deco(*a, **kw) 181 converted = convert_exception(e.java_exception) 182 if not isinstance(converted, UnknownException): 183 # Hide where the exception came from that shows a non-Pythonic 184 # JVM exception message. --> 185 raise converted from None 186 else: 187 raise IllegalArgumentException: requirement failed: source vector size 1 must be no less than k=2

from pyspark.ml.feature import PolynomialExpansion from pyspark.ml.linalg import Vectors data = [ (0, Vectors.dense([1.0, 2.0])), (1, Vectors.dense([2.0, 3.0])), (2, Vectors.dense([3.0, 4.0])) ] columns = ["id", "features"] df = spark.createDataFrame(data, columns) poly_expansion = PolynomialExpansion(inputCol="features", outputCol="expanded_features", degree=2) expanded_df = poly_expansion.transform(df) expanded_df.show(truncate=False)
+---+---------+-----------------------+ |id |features |expanded_features | +---+---------+-----------------------+ |0 |[1.0,2.0]|[1.0,1.0,2.0,2.0,4.0] | |1 |[2.0,3.0]|[2.0,4.0,3.0,6.0,9.0] | |2 |[3.0,4.0]|[3.0,9.0,4.0,12.0,16.0]| +---+---------+-----------------------+
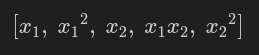



from pyspark.ml.feature import ChiSqSelector from pyspark.ml.linalg import Vectors data = [ (0, Vectors.dense([1.0, 0.1, -1.0]), 1.0), (1, Vectors.dense([2.0, 1.1, 1.0]), 0.0), (2, Vectors.dense([3.0, 10.1, 3.0]), 0.0) ] columns = ["id", "features", "label"] df = spark.createDataFrame(data, columns) selector = ChiSqSelector(numTopFeatures=1, featuresCol="features", outputCol="selected_features", labelCol="label") selector_model = selector.fit(df) selected_df = selector_model.transform(df) selected_df.show(truncate=False)
+---+--------------+-----+-----------------+ |id |features |label|selected_features| +---+--------------+-----+-----------------+ |0 |[1.0,0.1,-1.0]|1.0 |[1.0] | |1 |[2.0,1.1,1.0] |0.0 |[2.0] | |2 |[3.0,10.1,3.0]|0.0 |[3.0] | +---+--------------+-----+-----------------+
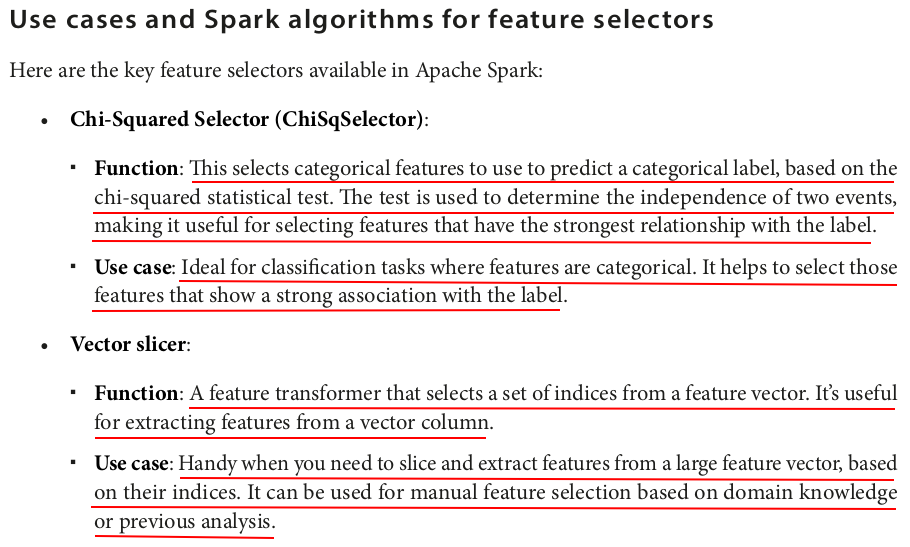
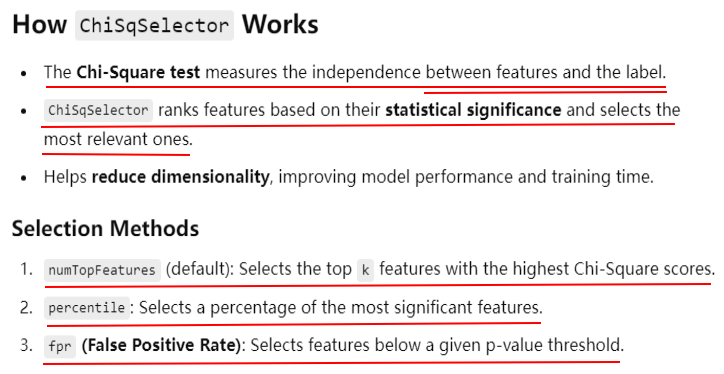
The ChiSqSelector selects the top k features based on the chi-squared test. In this example, we selected the single most important feature.

from pyspark.ml.feature import VectorSlicer from pyspark.ml.linalg import Vectors data = [ (0, Vectors.dense([1.0, 2.0, 3.0, 4.0, 5.0])), (1, Vectors.dense([2.0, 3.0, 4.0, 5.0, 6.0])), (2, Vectors.dense([3.0, 4.0, 5.0, 6.0, 7.0])) ] columns = ["id", "features"] df = spark.createDataFrame(data, columns) slicer = VectorSlicer(inputCol="features", outputCol="selected_features", indices=[1, 3, 4]) sliced_df = slicer.transform(df) sliced_df.show(truncate=False)
+---+---------------------+-----------------+ |id |features |selected_features| +---+---------------------+-----------------+ |0 |[1.0,2.0,3.0,4.0,5.0]|[2.0,4.0,5.0] | |1 |[2.0,3.0,4.0,5.0,6.0]|[3.0,5.0,6.0] | |2 |[3.0,4.0,5.0,6.0,7.0]|[4.0,6.0,7.0] | +---+---------------------+-----------------+

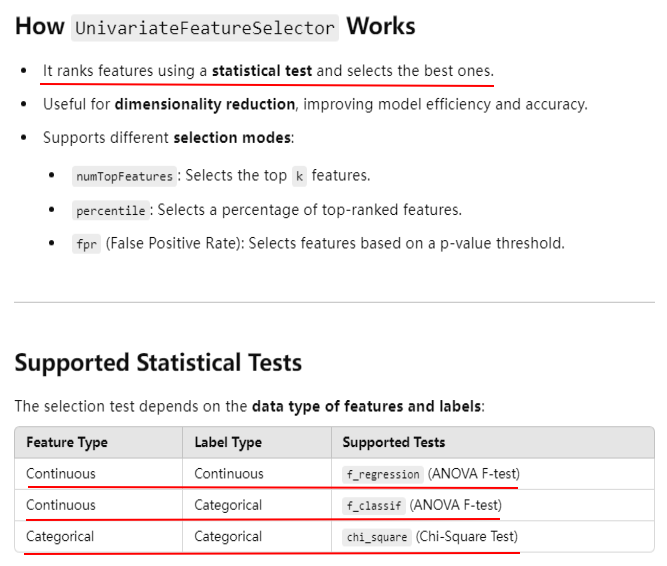
from pyspark.ml.feature import UnivariateFeatureSelector from pyspark.ml.linalg import Vectors data = [ (1.0, Vectors.dense([1.0, 0.1, -1.0])), (0.0, Vectors.dense([2.0, 1.1, 1.0])), (0.0, Vectors.dense([3.0, 10.1, 3.0])) ] columns = ["label", "features"] df = spark.createDataFrame(data, columns) selector = UnivariateFeatureSelector(featuresCol="features", outputCol="selected_features") selector.setFeatureType("continuous") \ .setLabelType("categorical") \ .setSelectionThreshold(1) selected_df = selector.fit(df).transform(df) selected_df.show(truncate=False)
+-----+--------------+-----------------+ |label|features |selected_features| +-----+--------------+-----------------+ |1.0 |[1.0,0.1,-1.0]|[1.0] | |0.0 |[2.0,1.1,1.0] |[2.0] | |0.0 |[3.0,10.1,3.0]|[3.0] | +-----+--------------+-----------------+





【推荐】国内首个AI IDE,深度理解中文开发场景,立即下载体验Trae
【推荐】编程新体验,更懂你的AI,立即体验豆包MarsCode编程助手
【推荐】抖音旗下AI助手豆包,你的智能百科全书,全免费不限次数
【推荐】轻量又高性能的 SSH 工具 IShell:AI 加持,快人一步
· 震惊!C++程序真的从main开始吗?99%的程序员都答错了
· 【硬核科普】Trae如何「偷看」你的代码?零基础破解AI编程运行原理
· 单元测试从入门到精通
· 上周热点回顾(3.3-3.9)
· winform 绘制太阳,地球,月球 运作规律