
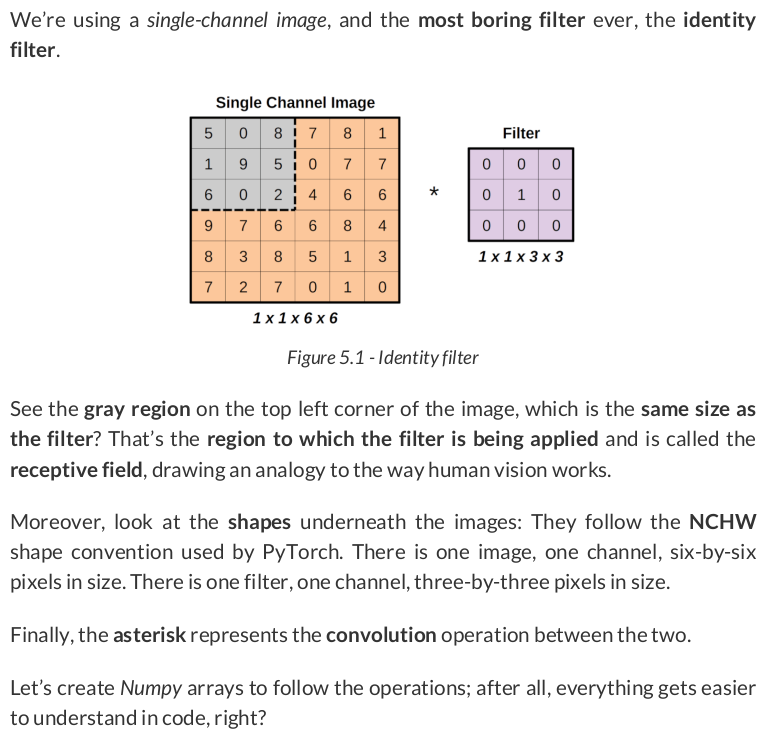
single = np.array( [[[[5, 0, 8, 7, 8, 1], [1, 9, 5, 0, 7, 7], [6, 0, 2, 4, 6, 6], [9, 7, 6, 6, 8, 4], [8, 3, 8, 5, 1, 3], [7, 2, 7, 0, 1, 0]]]] ) single.shape # (1, 1, 6, 6) identity = np.array( [[[[0, 0, 0], [0, 1, 0], [0, 0, 0]]]] ) identity.shape # (1, 1, 3, 3)
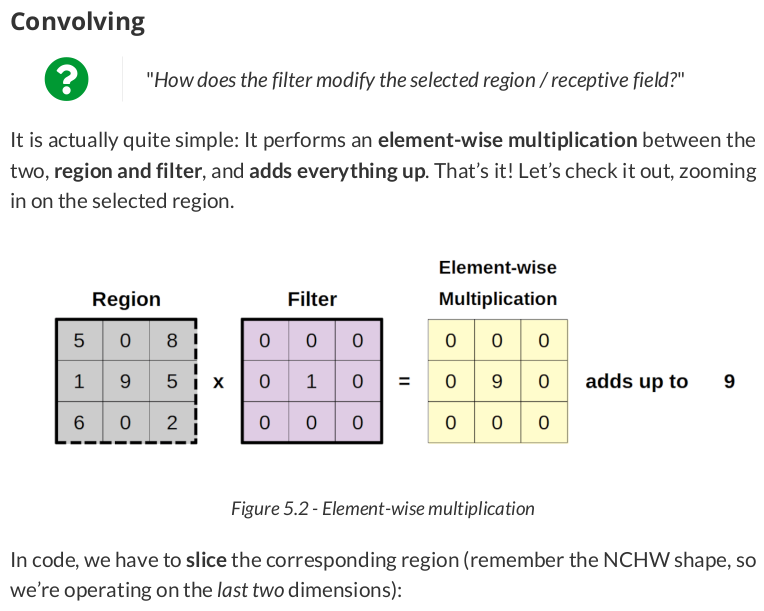
region = single[:, :, 0:3, 0:3] filtered_region = region * identity total = filtered_region.sum() total # np.int64(9)

new_region = single[:, :, 0:3, (0 + 1):(3 + 1)]
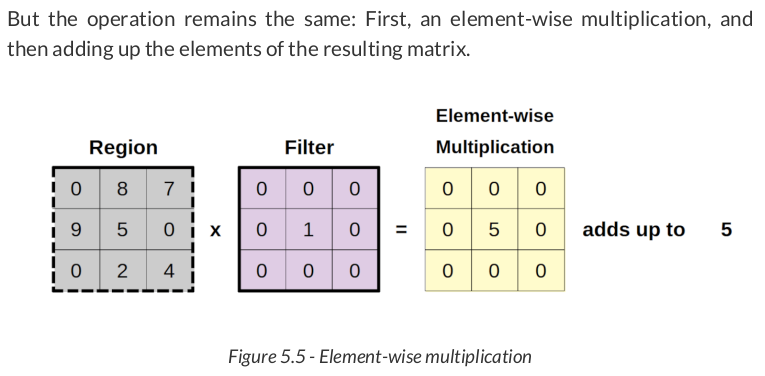
new_filtered_region = new_region * identity new_total = new_filtered_region.sum() new_total # np.int64(5)

last_horizontal_region = single[:, :, 0:3, (0 + 4):(3 + 4)]
The selected region does not match the shape of the filter anymore. So, if we try to perform the element-wise multiplication, it fails:
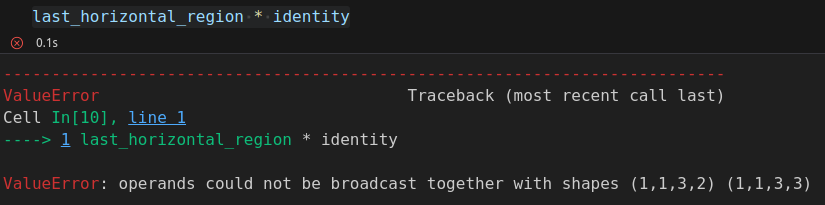
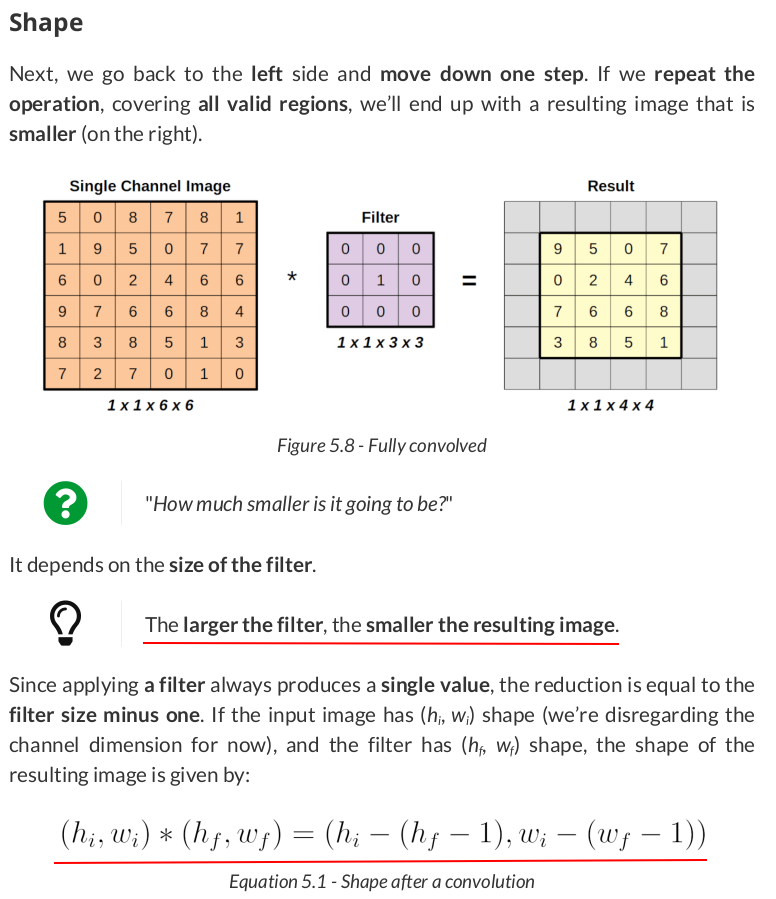
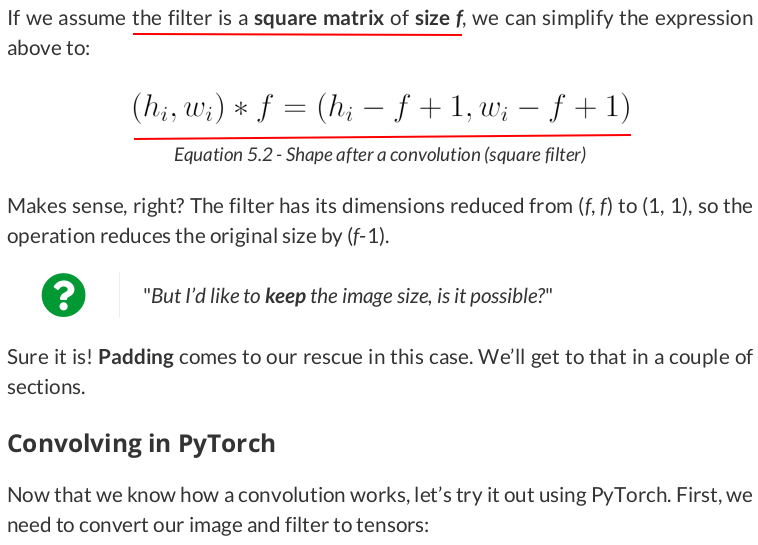
image = torch.as_tensor(single).float()
kernel_identity = torch.as_tensor(identity).float()
convolved = F.conv2d(image, kernel_identity, stride=1)
convolved
tensor([[[[9., 5., 0., 7.], [0., 2., 4., 6.], [7., 6., 6., 8.], [3., 8., 5., 1.]]]])

conv = nn.Conv2d(in_channels=1, out_channels=1, kernel_size=3, stride=1)
conv(image)
tensor([[[[-0.2127, 1.4696, 0.3466, -0.1149], [-1.8007, -0.3101, -3.0307, -2.1273], [-0.8260, -2.0202, -0.9840, 1.2278], [ 0.6377, 2.0343, 2.6733, 3.2409]]]], grad_fn=<ConvolutionBackward0>)

conv_multiple = nn.Conv2d(in_channels=1, out_channels=2, kernel_size=3, stride=1)
conv_multiple.weight
Parameter containing: tensor([[[[-0.0617, 0.2370, 0.3133], [ 0.1026, -0.3056, -0.2733], [-0.1523, -0.1558, 0.2972]]], [[[-0.1122, 0.2076, -0.0521], [-0.3075, -0.2325, -0.2017], [-0.2267, 0.0176, 0.1970]]]], requires_grad=True)

with torch.no_grad(): conv.weight[0] = kernel_identity conv.bias[0] = 0 conv(image)
tensor([[[[9., 5., 0., 7.], [0., 2., 4., 6.], [7., 6., 6., 8.], [3., 8., 5., 1.]]]], grad_fn=<ConvolutionBackward0>)

get the familiar result.



convolved_stride2 = F.conv2d(image, kernel_identity, stride=2) convolved_stride2 # tensor([[[[9., 0.], # [7., 6.]]]])


constant_padder = nn.ConstantPad2d(padding=1, value=0)
constant_padder(image)
constant_padder = nn.ConstantPad2d(padding=(1, 1, 1, 1), value=0)
constant_padder(image)
Same result:
tensor([[[[0., 0., 0., 0., 0., 0., 0., 0.],
[0., 5., 0., 8., 7., 8., 1., 0.],
[0., 1., 9., 5., 0., 7., 7., 0.],
[0., 6., 0., 2., 4., 6., 6., 0.],
[0., 9., 7., 6., 6., 8., 4., 0.],
[0., 8., 3., 8., 5., 1., 3., 0.],
[0., 7., 2., 7., 0., 1., 0., 0.],
[0., 0., 0., 0., 0., 0., 0., 0.]]]])

padded = F.pad(image, pad=(1, 1, 1, 1), value=0)
padded
padded = F.pad(image, pad=(1, 1, 1, 1), mode='constant', value=0) padded
Same result:
tensor([[[[0., 0., 0., 0., 0., 0., 0., 0.], [0., 5., 0., 8., 7., 8., 1., 0.], [0., 1., 9., 5., 0., 7., 7., 0.], [0., 6., 0., 2., 4., 6., 6., 0.], [0., 9., 7., 6., 6., 8., 4., 0.], [0., 8., 3., 8., 5., 1., 3., 0.], [0., 7., 2., 7., 0., 1., 0., 0.], [0., 0., 0., 0., 0., 0., 0., 0.]]]])


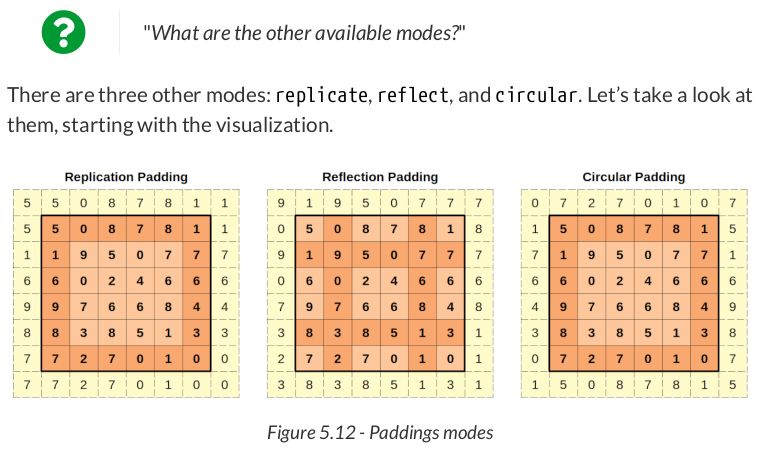

replication_padder = nn.ReplicationPad2d(padding=1)
replication_padder(image)
padded_replicate = F.pad(image, pad=(1, 1, 1, 1), mode='replicate') padded_replicate
Same result:
tensor([[[[5., 5., 0., 8., 7., 8., 1., 1.], [5., 5., 0., 8., 7., 8., 1., 1.], [1., 1., 9., 5., 0., 7., 7., 7.], [6., 6., 0., 2., 4., 6., 6., 6.], [9., 9., 7., 6., 6., 8., 4., 4.], [8., 8., 3., 8., 5., 1., 3., 3.], [7., 7., 2., 7., 0., 1., 0., 0.], [7., 7., 2., 7., 0., 1., 0., 0.]]]])

reflection_padder = nn.ReflectionPad2d(padding=1)
reflection_padder(image)
padded_reflect = F.pad(image, pad=(1, 1, 1, 1), mode='reflect') padded_reflect
Same result:
tensor([[[[9., 1., 9., 5., 0., 7., 7., 7.], [0., 5., 0., 8., 7., 8., 1., 8.], [9., 1., 9., 5., 0., 7., 7., 7.], [0., 6., 0., 2., 4., 6., 6., 6.], [7., 9., 7., 6., 6., 8., 4., 8.], [3., 8., 3., 8., 5., 1., 3., 1.], [2., 7., 2., 7., 0., 1., 0., 1.], [3., 8., 3., 8., 5., 1., 3., 1.]]]])
In PyTorch, you can use the functional form F.pad() with mode="reflect", or use the module version nn.ReflectionPad2d:
circular_padder = nn.CircularPad2d(padding=1)
circular_padder(image)
padded_circular = F.pad(image, pad=(1, 1, 1, 1), mode='circular') padded_circular
Same result:
tensor([[[[0., 7., 2., 7., 0., 1., 0., 7.], [1., 5., 0., 8., 7., 8., 1., 5.], [7., 1., 9., 5., 0., 7., 7., 1.], [6., 6., 0., 2., 4., 6., 6., 6.], [4., 9., 7., 6., 6., 8., 4., 9.], [3., 8., 3., 8., 5., 1., 3., 8.], [0., 7., 2., 7., 0., 1., 0., 7.], [1., 5., 0., 8., 7., 8., 1., 5.]]]])

edge = np.array( [[[[0, 1, 0], [1, -4, 1], [0, 1, 0]]]] ) kernel_edge = torch.as_tensor(edge).float() kernel_edge.shape # torch.Size([1, 1, 3, 3])
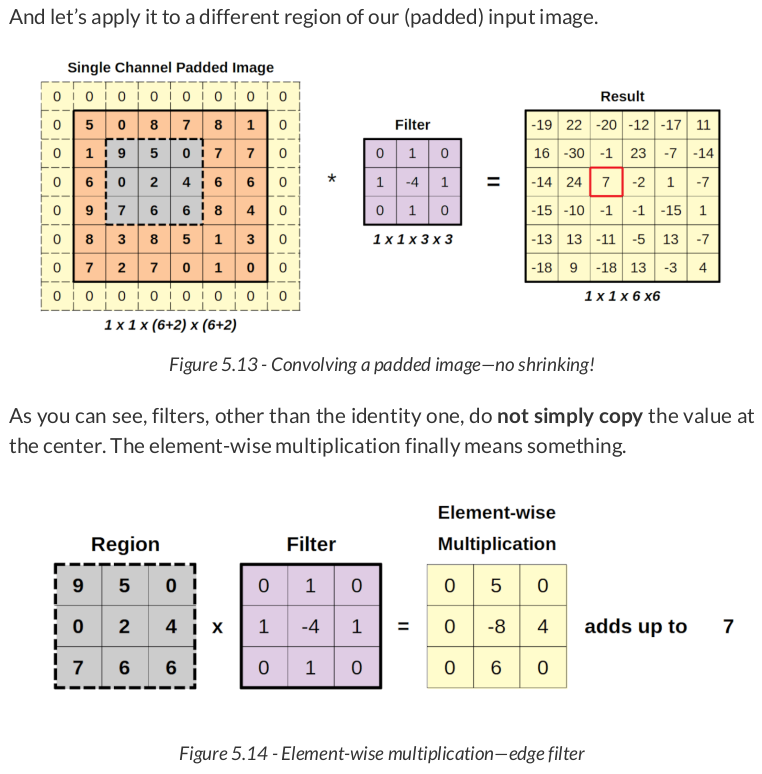
Let’s apply this filter to our image so we can use the resulting image in our next operation:
padded = F.pad(image, pad=(1, 1, 1, 1), mode='constant', value=0) conv_padded = F.conv2d(padded, kernel_edge, stride=1) conv_padded
tensor([[[[-19., 22., -20., -12., -17., 11.], [ 16., -30., -1., 23., -7., -14.], [-14., 24., 7., -2., 1., -7.], [-15., -10., -1., -1., -15., 1.], [-13., 13., -11., -5., 13., -7.], [-18., 9., -18., 13., -3., 4.]]]])
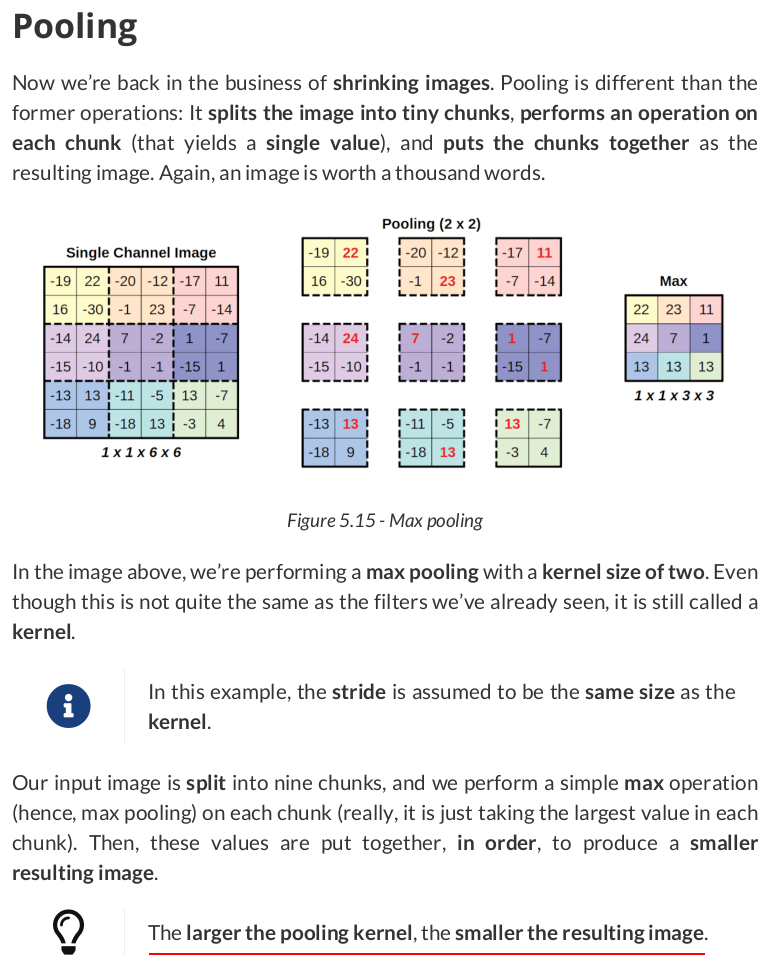

pooled = F.max_pool2d(conv_padded, kernel_size=2)
pooled
maxpooler = nn.MaxPool2d(kernel_size=2) pooled = maxpooler(conv_padded) pooled
Same result:
tensor([[[[22., 23., 11.], [24., 7., 1.], [13., 13., 13.]]]])
And then let’s use the module version to illustrate the large four-by-four pooling:
maxpooler4 = nn.MaxPool2d(kernel_size=4) pooled4 = maxpooler4(conv_padded) pooled4 # tensor([[[[24.]]]])


F.max_pool2d(conv_padded, kernel_size=3, stride=1)
tensor([[[[24., 24., 23., 23.], [24., 24., 23., 23.], [24., 24., 13., 13.], [13., 13., 13., 13.]]]])

flatenned = nn.Flatten()(pooled) flatenned # tensor([[22., 23., 11., 24., 7., 1., 13., 13., 13.]])
It has no functional version, but there is no need for one since we can accomplish the same thing using view():
pooled.view(1, -1) # tensor([[22., 23., 11., 24., 7., 1., 13., 13., 13.]])



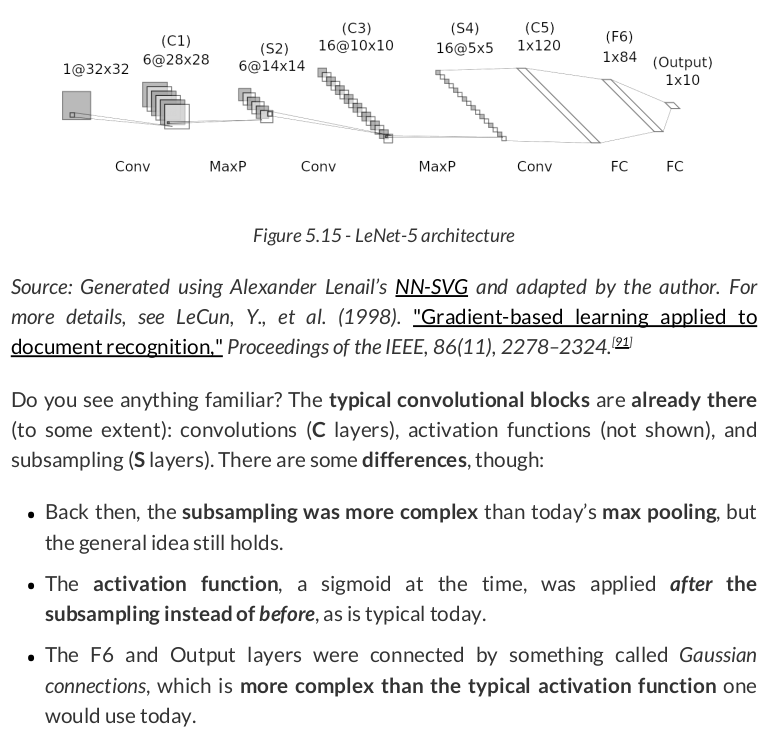
Adapting LeNet-5 to today’s standards, it could be implemented like this:
lenet = nn.Sequential() # Featurizer # Block 1: 1@28x28 -> 6@28x28 -> 6@14x14 lenet.add_module('C1', nn.Conv2d(in_channels=1, out_channels=6, kernel_size=5, padding=2)) lenet.add_module('func1', nn.ReLU()) lenet.add_module('S2', nn.MaxPool2d(kernel_size=2)) # Block 2: 6@14x14 -> 16@10x10 -> 16@5x5 lenet.add_module('C3', nn.Conv2d(in_channels=6, out_channels=16, kernel_size=5)) lenet.add_module('func2', nn.ReLU()) lenet.add_module('S4', nn.MaxPool2d(kernel_size=2)) # Block 3: 16@5x5 -> 120@1x1 lenet.add_module('C5', nn.Conv2d(in_channels=15, out_channels=120, kernel_size=5)) lenet.add_module('func2', nn.ReLU()) # Flattening lenet.add_module('flatten', nn.Flatten()) # Classification # Hidden Layer lenet.add_module('F6', nn.Linear(in_features=120, out_features=84)) lenet.add_module('func3', nn.ReLU()) # Output Layer lenet.add_module('OUTPUT', nn.Linear(in_features=84, out_features=10))



images, labels = generate_dataset(img_size=10, n_images=1000, binary=False, seed=17)
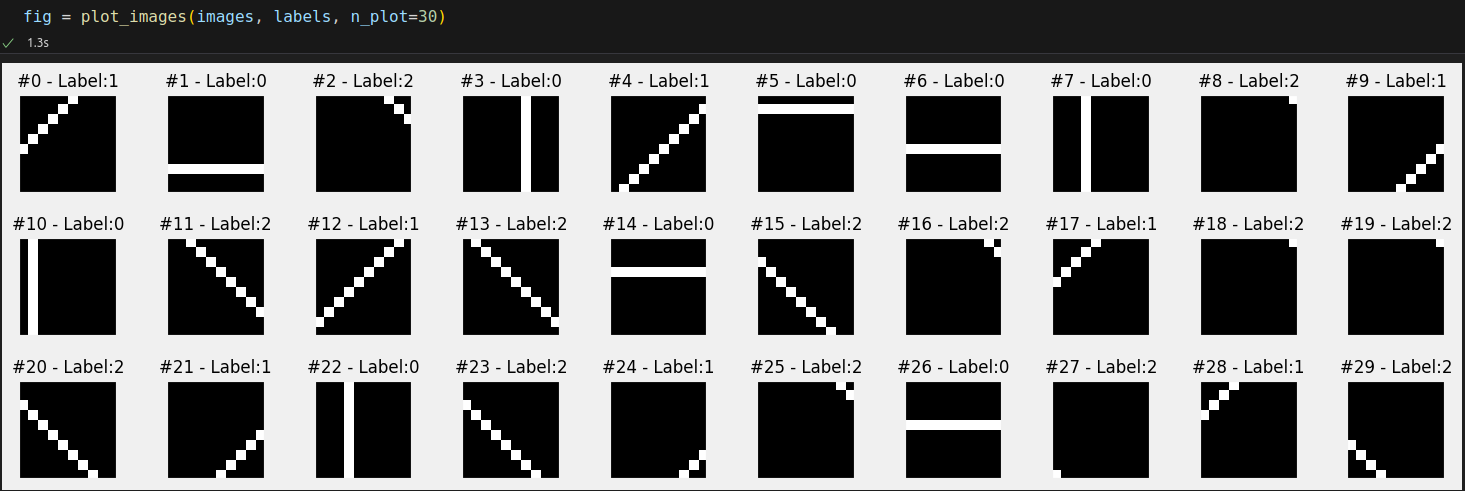
# Build tensors from numpy arrays BEFORE split # Modify the scale of pixel values from [0, 255] to [0, 1] x_tensor = torch.as_tensor(images / 255).float() y_tensor = torch.as_tensor(labels).long() # Use index_splitter to generate indices for training and validation sets train_idx, val_idx = index_splitter(len(x_tensor), [80, 20]) # Use indices to perform the split x_train_tensor = x_tensor[train_idx] y_train_tensor = y_tensor[train_idx] x_val_tensor = x_tensor[val_idx] y_val_tensor = y_tensor[val_idx] # We're not doing any data augmentation now train_composer = Compose([Normalize(mean=(.5,), std=(.5,))]) val_composer = Compose([Normalize(mean=(.5,), std=(.5,))]) # Use custom dataset to apply composed transforms to each set train_dataset = TransformedTensorDataset(x_train_tensor, y_train_tensor, transform=train_composer) val_dataset = TransformedTensorDataset(x_val_tensor, y_val_tensor, transform=val_composer) # Build a weighted random sampler to handle imbalanced classes sampler = make_balanced_sampler(y_train_tensor) # Use sampler in the training set to get a balanced data loader train_loader = DataLoader(dataset=train_dataset, batch_size=16, sampler=sampler) val_loader = DataLoader(dataset=val_dataset, batch_size=16)
Before defining a model to classify our images, we need to discuss something else: the loss function.


logits = torch.tensor([ 1.3863, 0.0000, -0.6931])
We exponentiate the logits to get the corresponding odds ratios:
odds_ratios = torch.exp(logits) odds_ratios # tensor([4.0000, 1.0000, 0.5000])

softmaxed = odds_ratios / odds_ratios.sum() softmaxed # tensor([0.7273, 0.1818, 0.0909])

nn.Softmax(dim=-1)(logits), F.softmax(logits, dim=-1) # (tensor([0.7273, 0.1818, 0.0909]), tensor([0.7273, 0.1818, 0.0909]))



logits = torch.tensor([ 1.3863, 0.0000, -0.6931]) log_probs = F.log_softmax(logits, dim=-1) log_probs # tensor([-0.3185, -1.7048, -2.3979])

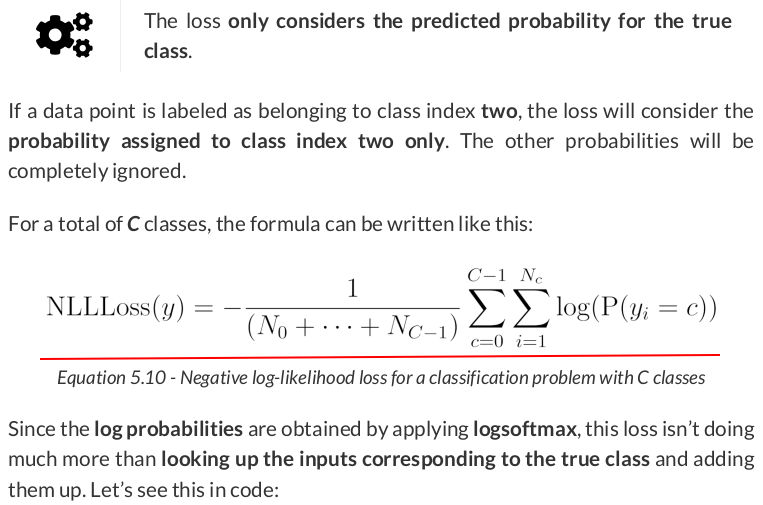
label = torch.tensor([2]) F.nll_loss(log_probs.view(-1, 3), label) # tensor(2.3979)
It is the negative of the log probability corresponding to the class index (two) of the true label.

torch.manual_seed(11) dummy_logits = torch.randn((5, 3)) dummy_labels = torch.tensor([0, 0, 1, 2, 1]) dummy_log_probs = F.log_softmax(dummy_logits, dim=-1) dummy_log_probs
tensor([[-1.5229, -0.3146, -2.9600], [-1.7934, -1.0044, -0.7607], [-1.2513, -1.0136, -1.0471], [-2.6799, -0.2219, -2.0367], [-1.0728, -1.9098, -0.6737]])

relevant_log_probs = torch.tensor([-1.5229, -1.7934, -1.0136, -2.0367, -1.9098]) -relevant_log_probs.mean() # tensor(1.6553)

loss_fn = nn.NLLLoss() loss_fn(dummy_log_probs, dummy_labels) # tensor(1.6553)

loss_fn = nn.NLLLoss(weight=torch.tensor([1., 1., 2.])) loss_fn(dummy_log_probs, dummy_labels) # tensor(1.7188)

loss_fn = nn.NLLLoss(ignore_index=2) loss_fn(dummy_log_probs, dummy_labels) # tensor(1.5599)


torch.manual_seed(11) dummy_logits = torch.randn((5, 3)) dummy_labels = torch.tensor([0, 0, 1, 2, 1]) loss_fn = nn.CrossEntropyLoss() loss_fn(dummy_logits, dummy_labels) # tensor(1.6553)
No logsoftmax whatsoever, but the same resulting loss, as expected.


torch.manual_seed(13) model_cnn1 = nn.Sequential() # Featurizer # Block 1: 1@10x10 -> n_channels@8x8 -> n_channels@4x4 n_channels = 1 model_cnn1.add_module('conv1', nn.Conv2d(in_channels=1, out_channels=n_channels, kernel_size=3)) model_cnn1.add_module('relu1', nn.ReLU()) model_cnn1.add_module('maxp1', nn.MaxPool2d(kernel_size=2)) # Flattening: n_channels * 4 * 4 model_cnn1.add_module('flatten', nn.Flatten())



lr = .1 ce_loss_fn = nn.CrossEntropyLoss(reduction='mean') optimizer_cnn1 = optim.SGD(model_cnn1.parameters(), lr=lr)

sbs_cnn1 = StepByStep(model_cnn1, ce_loss_fn, optimizer_cnn1)
sbs_cnn1.set_loaders(train_loader, val_loader)
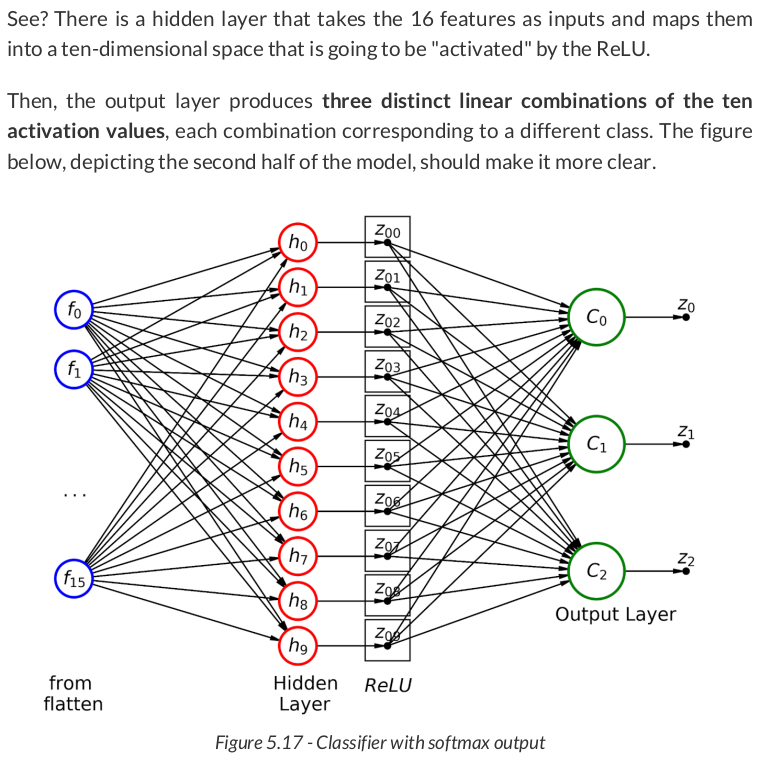
sbs_cnn1.train(20)

@staticmethod def _visualize_tensors(axs, x, y=None, yhat=None, layer_name='', title=None): n_images = len(axs) # Gets max and min values for scaling the grayscale minv, maxv = np.min(x[:n_images]), np.max(x[:n_images]) for i, img in enumerate(x[:n_images]): ax = axs[i] # Set title, labels, and remove ticks if title is not None: ax.set_title(f'{title} #{i}', fontsize=12) shp = np.atleast_2d(img).shape ax.set_ylabel(f'{layer_name}\n{shp[0]}x{shp[1]}', rotation=0, fontsize=12, labelpad=20) xlabel1 = '' if y is None else f'\nLabel: {y[i]}' xlabel2 = '' if yhat is None else f'\nPredicted: {yhat[i]}' xlabel = f'{xlabel1}{xlabel2}' if len(xlabel): ax.set_xlabel(xlabel, fontsize=12) ax.set_xticks([]) ax.set_yticks([]) # Plot weight as an image ax.imshow(np.atleast_2d(img.squeeze()), cmap='gray', vmin=minv, vmax=maxv) return

weights_filter = model_cnn1.conv1.weight.data.cpu().numpy() weights_filter.shape # (1, 1, 3, 3)

def visualize_filters(self, layer_name, **kwargs): try: # Get the layer object from the model layer = getattr(self.model, layer_name) # We are only looking at filters for 2D convolutions if isinstance(layer, nn.Conv2d): weights = layer.weight.data.cpu().numpy() # weights -> (ou_channels (filter), in_channels, filter_H, filter_W) n_filters, n_in_channels, _, _ = weights.shape # Build a figure figsize = (2 * n_in_channels + 2, 2 * n_filters) fig, axs = plt.subplots(n_filters, n_in_channels, figsize=figsize, squeeze=False) axs_array = [[axs[i, j] for j in range(n_in_channels)] for i in range(n_filters)] # For each filter for i in range(n_filters): StepByStep._visualize_tensors( axs_array[i], weights[i], layer_name=f'Filter #{i}', title='Channel' ) for ax in axs.flat: ax.label_outer() fig.tight_layout() return except AttributeError: return
OK, let’s see what the filter looks like:

dummy_model = nn.Linear(1, 1) dummy_list = [] def dummy_hook(layer, inputs, outputs): dummy_list.append((layer, inputs, outputs))

dummy_handle = dummy_model.register_forward_hook(dummy_hook) dummy_handle # <torch.utils.hooks.RemovableHandle at 0x727670b65eb0>
Simple enough, right? Let’s see it in action:
dummy_x = torch.tensor([0.3]) dummy_model.forward(dummy_x) # tensor([-0.8366], grad_fn=<ViewBackward0>)

dummy_list # []

dummy_model(dummy_x) # tensor([-0.8366], grad_fn=<ViewBackward0>)
dummy_list # [(Linear(in_features=1, out_features=1, bias=True), # (tensor([0.3000]),), # tensor([-0.8366], grad_fn=<ViewBackward0>))]

dummy_handle.remove()

modules = list(sbs_cnn1.model.named_modules())
modules
[('', Sequential( (conv1): Conv2d(1, 1, kernel_size=(3, 3), stride=(1, 1)) (relu1): ReLU() (maxp1): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False) (flatten): Flatten(start_dim=1, end_dim=-1) (fc1): Linear(in_features=16, out_features=10, bias=True) (relu2): ReLU() (fc2): Linear(in_features=10, out_features=3, bias=True) )), ('conv1', Conv2d(1, 1, kernel_size=(3, 3), stride=(1, 1))), ('relu1', ReLU()), ('maxp1', MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)), ('flatten', Flatten(start_dim=1, end_dim=-1)), ('fc1', Linear(in_features=16, out_features=10, bias=True)), ('relu2', ReLU()), ('fc2', Linear(in_features=10, out_features=3, bias=True))]

layer_names = {layer: name for name, layer in modules[1:]}
layer_names
{Conv2d(1, 1, kernel_size=(3, 3), stride=(1, 1)): 'conv1',
ReLU(): 'relu1',
MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False): 'maxp1',
Flatten(start_dim=1, end_dim=-1): 'flatten',
Linear(in_features=16, out_features=10, bias=True): 'fc1',
ReLU(): 'relu2',
Linear(in_features=10, out_features=3, bias=True): 'fc2'}

visualization = {} def hook_fn(layer, inputs, outputs): name = layer_names[layer] visualization[name] = outputs.detach().cpu().numpy()

layers_to_hook = ['conv1', 'relu1', 'maxp1', 'flatten', 'fc1', 'relu2', 'fc2'] handles = {} for name, layer in modules: if name in layers_to_hook: handles[name] = layer.register_forward_hook(hook_fn)

images_batch, labels_batch = next(iter(val_loader))
logits = sbs_cnn1.predict(images_batch)
Now, if everything went well, our visualization dictionary should contain one key for each layer we hooked a function to:
visualization.keys() # dict_keys(['conv1', 'relu1', 'maxp1', 'flatten', 'fc1', 'relu2', 'fc2'])
Bingo! They are all there! But, before checking what’s stored inside it, let’s remove the hooks:
for handle in handles.values(): handle.remove() handles
{'conv1': <torch.utils.hooks.RemovableHandle at 0x727671e38860>,
'relu1': <torch.utils.hooks.RemovableHandle at 0x727671f16540>,
'maxp1': <torch.utils.hooks.RemovableHandle at 0x727671f73320>,
'flatten': <torch.utils.hooks.RemovableHandle at 0x727671f71940>,
'fc1': <torch.utils.hooks.RemovableHandle at 0x727671f72870>,
'relu2': <torch.utils.hooks.RemovableHandle at 0x727671f73fe0>,
'fc2': <torch.utils.hooks.RemovableHandle at 0x727671f16e70>}


Update class StepByStep:
self.visualization = {} self.handles = {} def attach_hooks(self, layers_to_hook, hook_fn=None): # Clear any previous values self.visualization = {} # Create the dictionary to map layer objects to their names modules = list(self.model.named_modules()) layer_names = {layer: name for name, layer in modules[1:]} if hook_fn is None: # Hook function to be attached to the forward pass def hook_fn(layer, inputs, outputs): # Get the layer name name = layer_names[layer] # Detach the outputs values = outputs.detach().cpu().numpy() # Since the hook function may be called multiple times for example, # if we make predictions for multiple mini-batches it concatenates the results. if self.visualization[name] is None: self.visualization[name] = values else: self.visualization[name] = np.concatenate([self.visualization[name], values]) for name, layer in modules: if name in layers_to_hook: # Initialize the corresponding key in the dictionary self.visualization[name] = None # Register the forward hook and keep the handle in another dict self.handles[name] = layer.register_forward_hook(hook_fn) def remove_hooks(self): for handle in self.handles.values(): handle.remove() # Clear the dict, as all hooks have been removed self.handles = {}
The procedure is fairly straightforward now: Give it a list containing the names of the layers to attach hooks to, and you’re done!
sbs_cnn1.attach_hooks(layers_to_hook=['conv1', 'relu1', 'maxp1', 'flatten', 'fc1', 'relu2', 'fc2'])
To get the visualization attribute filled with values, we still need to make predictions:
images_batch, labels_batch = next(iter(val_loader)) logits = sbs_cnn1.predict(images_batch) logits.shape # (16, 3)

sbs_cnn1.remove_hooks()

predicted = np.argmax(logits, axis=1) predicted # array([0, 0, 0, 2, 2, 1, 0, 2, 1, 2, 0, 1, 0, 2, 2, 1])

First, let’s visualize the first ten images sampled from the validation loader:


for layer, feature_map in sbs_cnn1.visualization.items(): print(layer, feature_map.shape)
conv1 (16, 1, 8, 8) relu1 (16, 1, 8, 8) maxp1 (16, 1, 4, 4) flatten (16, 16) fc1 (16, 10) relu2 (16, 10) fc2 (16, 3)
To visualize the feature maps, we can add another method to our class: visualize_outputs(). This method simply retrieves the captured feature maps from the visualization dictionary and uses our _visualize_tensors() method to plot them:
def visualize_outputs(self, layers, n_images=16, y=None, yhat=None): layers = filter(lambda l: l in self.visualization.keys(), layers) layers = list(layers) shapes = [self.visualization[layer].shape for layer in layers] n_rows = [shape[1] if len(shape) == 4 else 1 for shape in shapes] # number of output channels total_rows = np.sum(n_rows) fig, axs = plt.subplots(total_rows, n_images, figsize=(1.5 * n_images, 1.5 * total_rows), squeeze=False) axs_array = [[axs[i, j] for j in range(n_images)] for i in range(total_rows)] # Loop through the layers, one layer per row of subplots row = 0 for i, layer in enumerate(layers): start_row = row # Take the produced feature maps for that layer output = self.visualization[layer] is_vector = len(output.shape) == 2 for j in range(n_rows[i]): StepByStep._visualize_tensors( axs_array[row], output if is_vector else output[:, j].squeeze(), y, yhat, layer_name=layers[i] if is_vector else f'{layers[i]}\nfil#{row - start_row}', title='Image' if row == 0 else None ) row += 1 for ax in axs.flat: ax.label_outer() fig.tight_layout() return fig
Then, let’s use the method above to plot the feature maps for the layers in the featurizer part of our model:





• For diagonals tilted to the left (images #3, #4, #7, #13 and #14), the filter seems to suppress the diagonal completely.
• For parallel lines (images #0, #1, #2, #6, #9, #10, and #12), the filter produces a striped pattern, brighter to the left/top of the original line, darker to its right/bottom.
• For diagonals tilted to the right (images #11 and #15), the filter produces a thicker line with multiple shades.
Then, the ReLU activation function removes the negative values. Unfortunately, after this operation, image #9 (parallel horizontal lines) had all lines suppressed and seem indistinguishable from images #3, #4, #7, # 13 and #14 (diagonals tilted to the left).
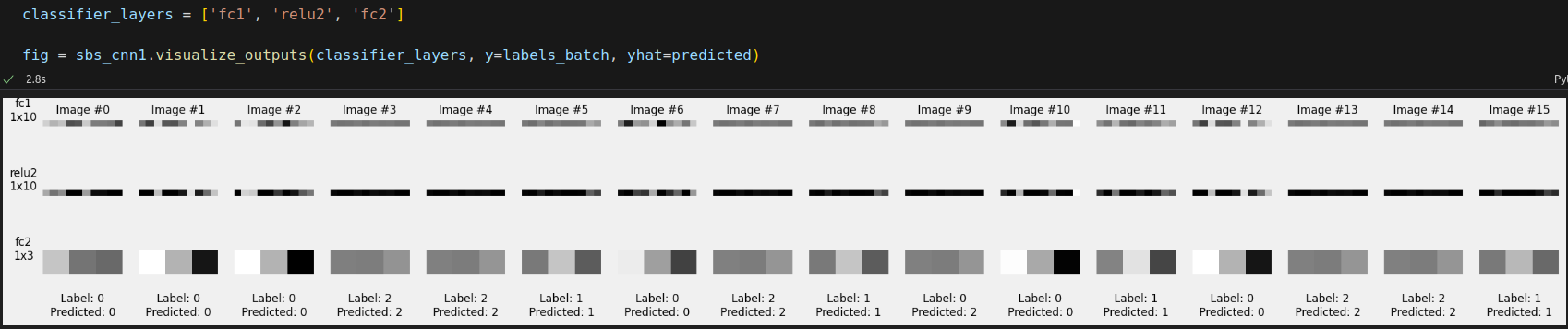

The classifier got 15 out of 16 right. It made wrong prediction for image #9. Unsurprisingly, this is the image that got its horizontal line suppressed. The filter doesn’t seem to work so well whenever the horizontal line is too close to the top edge of the image.
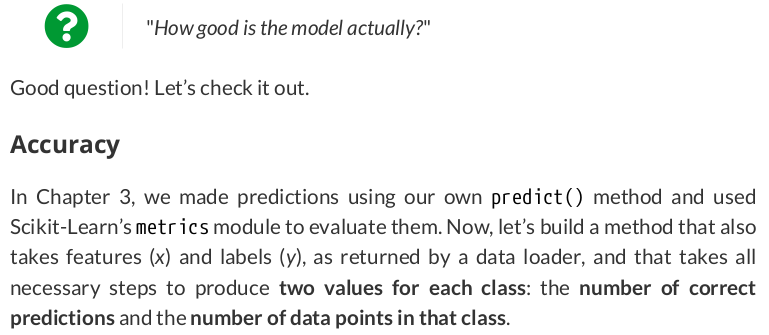
def correct(self, x, y, threshold=.5): self.model.eval() yhat = self.model(x.to(self.device)) y = y.to(self.device) self.model.train() # We get the size of the batch and the number of classes (only 1, if it is binary) n_samples, n_dims = yhat.shape if n_dims > 1: # In a multiclass classification, the biggest logit always wins, so we don't bother getting probabilities # This is PyTorch's version of argmax, but it returns a tuple: (max value, index of max value) _, predicted = torch.max(yhat, 1) else: n_dims += 1 # In binary classification, we NEED to check if the last layer is a sigmoid (and then it produces probs) if isinstance(self.model, nn.Sequential) and isinstance(self.model[-1], nn.Sigmoid): predicted = (yhat > threshold).long() # or something else (logits), which we need to convert using a sigmoid else: predicted = (F.sigmoid(yhat) > threshold).long() # How many samples got classified correctly for each class result = [] for c in range(n_dims): n_class = (y == c).sum().item() n_correct = (predicted[y == c] == c).sum().item() result.append((n_correct, n_class)) return torch.tensor(result) setattr(StepByStep, 'correct', correct)
If the labels have two or more columns, it means we’re dealing with a multiclass classification: The predicted class is the one with the largest logit.
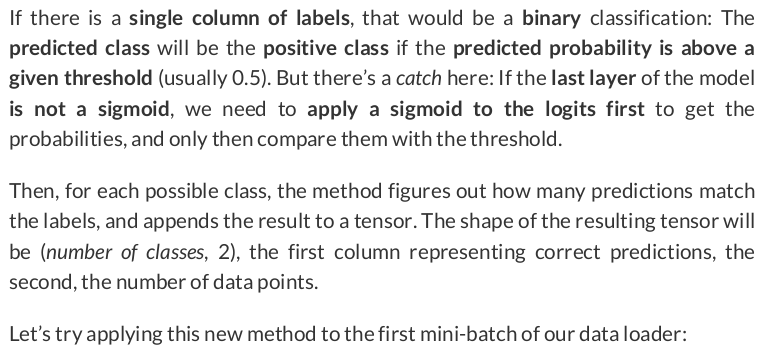
sbs_cnn1.correct(images_batch, labels_batch)
tensor([[6, 7], [4, 4], [5, 5]])
So, there is only 1 wrong prediction, for class #0 (parallel lines), corresponding to image #9, as we’ve already seen in the previous section.

@staticmethod def loader_apply(loader, func, reduce='sum'): results = [func(x, y) for i, (x, y) in enumerate(loader)] results = torch.stack(results, axis=0) if reduce == 'sum': results = results.sum(axis=0) elif reduce == 'mean': results = results.float().mean(axis=0) return results setattr(StepByStep, 'loader_apply', loader_apply)

StepByStep.loader_apply(sbs_cnn1.val_loader, sbs_cnn1.correct)
tensor([[47, 58], [57, 67], [75, 75]])

From the results above, we see that our model got 179 out of 200 images correctly classified in the validation set, an accuracy of 89.5%! Not bad :-)


Data Preparation
# Build tensors from numpy arrays BEFORE split # Modify the scale of pixel values from [0, 255] to [0, 1] x_tensor = torch.as_tensor(images / 255).float() y_tensor = torch.as_tensor(labels).long() # Use index_splitter to generate indices for training and validation sets train_idx, val_idx = index_splitter(len(x_tensor), [80, 20]) # Use indices to perform the split x_train_tensor = x_tensor[train_idx] y_train_tensor = y_tensor[train_idx] x_val_tensor = x_tensor[val_idx] y_val_tensor = y_tensor[val_idx] # We're not doing any data augmentation now train_composer = Compose([Normalize(mean=(.5,), std=(.5,))]) val_composer = Compose([Normalize(mean=(.5,), std=(.5,))]) # Use custom dataset to apply composed transforms to each set train_dataset = TransformedTensorDataset(x_train_tensor, y_train_tensor, transform=train_composer) val_dataset = TransformedTensorDataset(x_val_tensor, y_val_tensor, transform=val_composer) # Build a weighted random sampler to handle imbalanced classes sampler = make_balanced_sampler(y_train_tensor) # Use sampler in the training set to get a balanced data loader train_loader = DataLoader(dataset=train_dataset, batch_size=16, sampler=sampler) val_loader = DataLoader(dataset=val_dataset, batch_size=16)
Model Configuration
torch.manual_seed(13) model_cnn1 = nn.Sequential() # Featurizer # Block 1: 1@10x10 -> n_channels@8x8 -> n_channels@4x4 n_channels = 1 model_cnn1.add_module('conv1', nn.Conv2d(in_channels=1, out_channels=n_channels, kernel_size=3)) model_cnn1.add_module('relu1', nn.ReLU()) model_cnn1.add_module('maxp1', nn.MaxPool2d(kernel_size=2)) # Flattening: n_channels * 4 * 4 model_cnn1.add_module('flatten', nn.Flatten()) # Classification # Hidden Layer model_cnn1.add_module('fc1', nn.Linear(in_features=n_channels*4*4, out_features=10)) model_cnn1.add_module('relu2', nn.ReLU()) # Output Layer model_cnn1.add_module('fc2', nn.Linear(in_features=10, out_features=3)) lr = .1 ce_loss_fn = nn.CrossEntropyLoss(reduction='mean') optimizer_cnn1 = optim.SGD(model_cnn1.parameters(), lr=lr)
Model Training
sbs_cnn1 = StepByStep(model_cnn1, ce_loss_fn, optimizer_cnn1)
sbs_cnn1.set_loaders(train_loader, val_loader)
sbs_cnn1.train(20)
Visualizing Filters
fig_filters = sbs_cnn1.visualize_filters('conv1', cmap='gray')
Capturing Outputs
featurizer_layers = ['conv1', 'relu1', 'maxp1', 'flatten'] classifier_layers = ['fc1', 'relu2', 'fc2'] sbs_cnn1.attach_hooks(layers_to_hook=featurizer_layers + classifier_layers) images_batch, labels_batch = next(iter(val_loader)) logits = sbs_cnn1.predict(images_batch) predicted = np.argmax(logits, 1) sbs_cnn1.remove_hooks()
Visualizing Feature Maps
fig_maps1 = sbs_cnn1.visualize_outputs(featurizer_layers)
fig_maps2 = sbs_cnn1.visualize_outputs(classifier_layers, y=labels_batch, yhat=predicted)
Evaluating Accuracy
StepByStep.loader_apply(sbs_cnn1.val_loader, sbs_cnn1.correct)


 浙公网安备 33010602011771号
浙公网安备 33010602011771号