逻辑回归Stepwise的R代码
逻辑回归见前述随笔。缩进全乱了。
1

 pi_fun<-function(x,Beta)
pi_fun<-function(x,Beta) {
{2
 Beta<-matrix(as.vector(Beta),ncol=1)
Beta<-matrix(as.vector(Beta),ncol=1)3
 x<-matrix(as.vector(x),ncol=nrow(Beta))
x<-matrix(as.vector(x),ncol=nrow(Beta))4
 g_fun<-x%*%Beta
g_fun<-x%*%Beta 5
 exp(g_fun)/(1+exp(g_fun))
exp(g_fun)/(1+exp(g_fun)) 6
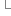 }
} 7

8

 funs<-function(x,y,Beta)
funs<-function(x,y,Beta) {
{9
 y<-matrix(as.vector(y),ncol=1)
y<-matrix(as.vector(y),ncol=1)10
 x<-matrix(as.vector(x),nrow=nrow(y))
x<-matrix(as.vector(x),nrow=nrow(y))11
 Beta<-matrix(c(Beta),ncol=1)
Beta<-matrix(c(Beta),ncol=1)12

13
 pi_value<- pi_fun(x,Beta)
pi_value<- pi_fun(x,Beta)14
 U<-t(x)%*%(y-pi_value);
U<-t(x)%*%(y-pi_value);15
 uni_matrix<-matrix(rep(1,nrow(pi_value)),nrow= nrow(pi_value));
uni_matrix<-matrix(rep(1,nrow(pi_value)),nrow= nrow(pi_value));16
 H<-t(x)%*%diag(as.vector(pi_value*( uni_matrix -pi_value)))%*%x
H<-t(x)%*%diag(as.vector(pi_value*( uni_matrix -pi_value)))%*%x17
 list(U=U,H=H)
list(U=U,H=H)18
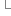 }
}19

20

 Newtons<-function(fun,x,y,ep=1e-8,it_max=100)
Newtons<-function(fun,x,y,ep=1e-8,it_max=100) {
{21
 x<-matrix(as.vector(x),nrow=nrow(y))
x<-matrix(as.vector(x),nrow=nrow(y))22
 Beta=matrix(rep(0,ncol(x)),nrow=ncol(x))
Beta=matrix(rep(0,ncol(x)),nrow=ncol(x))23
 Index<-0;
Index<-0;24
 k<-1
k<-125

 while(k<=it_max)
while(k<=it_max) {
{26
 x1<-Beta;obj<-fun(x,y,Beta);
x1<-Beta;obj<-fun(x,y,Beta);27

28
 Beta<-Beta+solve(obj$H,obj$U);
Beta<-Beta+solve(obj$H,obj$U);29

30
 objTemp<-fun(x,y,Beta)
objTemp<-fun(x,y,Beta)31

 if(any(is.nan(objTemp$H)))
if(any(is.nan(objTemp$H))) {
{32
 Beta<-x1
Beta<-x133
 print("Warning:The maximum likelihood estimate does not exist.")
print("Warning:The maximum likelihood estimate does not exist.")34
 print(paste("The LOGISTIC procedure continues in spite of the above warning.",
print(paste("The LOGISTIC procedure continues in spite of the above warning.",35
 "Results shown are based on the last maximum likelihood iteration. Validity of the model fit is questionable."));
"Results shown are based on the last maximum likelihood iteration. Validity of the model fit is questionable."));36
 break
break37
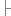 }
} 38
 norm<-sqrt(t((Beta-x1))%*%(Beta-x1))
norm<-sqrt(t((Beta-x1))%*%(Beta-x1))39

 if(norm<ep)
if(norm<ep) {
{40
 index<-1;break
index<-1;break41
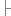 }
}42
 k<-k+1
k<-k+143
 }
}44
 obj<-fun(x,y,Beta);
obj<-fun(x,y,Beta);45

46
 list(Beta=Beta,it=k,U=obj$U,H=obj$H)
list(Beta=Beta,it=k,U=obj$U,H=obj$H)47
 }
}48

49

 CheckZero<-function(x)
CheckZero<-function(x) {
{50

 for(i in 1:length(x))
for(i in 1:length(x)) {
{51
 if(x[i]==0)
if(x[i]==0)52
 x[i]<-1e-300
x[i]<-1e-30053
 }
} 54
 x
x55
 }
} 56

57

 ModelFitStat<-function(x,y,Beta)
ModelFitStat<-function(x,y,Beta) {
{58
 x<-matrix(as.vector(x),nrow=nrow(y))
x<-matrix(as.vector(x),nrow=nrow(y))59
 Beta<-matrix(as.vector(Beta),ncol=1)
Beta<-matrix(as.vector(Beta),ncol=1)60

61
 uni_matrix<-matrix(rep(1,nrow(y)),nrow= nrow(y));
uni_matrix<-matrix(rep(1,nrow(y)),nrow= nrow(y));62
 LOGL<--2*(t(y)%*%log(pi_fun(x,Beta))+t(uni_matrix-y)%*%log(CheckZero(uni_matrix-pi_fun(x,Beta))))
LOGL<--2*(t(y)%*%log(pi_fun(x,Beta))+t(uni_matrix-y)%*%log(CheckZero(uni_matrix-pi_fun(x,Beta))))63

64
 #print("-----------------")
#print("-----------------")65
 #print(LOGL)
#print(LOGL)66

67
 AIC<-LOGL+2*(ncol(x))
AIC<-LOGL+2*(ncol(x))68
 SC<-LOGL+(ncol(x))*log(nrow(x))
SC<-LOGL+(ncol(x))*log(nrow(x))69
 list(LOGL=LOGL,AIC=AIC,SC=SC)
list(LOGL=LOGL,AIC=AIC,SC=SC)70

71
 }
}72

73

74

 GlobalNullTest<-function(x,y,Beta,BetaIntercept)
GlobalNullTest<-function(x,y,Beta,BetaIntercept) {
{75
 y<-matrix(as.vector(y),ncol=1)
y<-matrix(as.vector(y),ncol=1)76
 x<-matrix(as.vector(x),nrow=nrow(y))
x<-matrix(as.vector(x),nrow=nrow(y))77
 Beta<-matrix(as.vector(Beta),ncol=1)
Beta<-matrix(as.vector(Beta),ncol=1)78

79
 pi_value<- pi_fun(x,Beta)
pi_value<- pi_fun(x,Beta)80
 df<-nrow(Beta)-1
df<-nrow(Beta)-181
 ##compute Likelihood ratio
##compute Likelihood ratio82

83
 MF<-ModelFitStat(x,y,Beta)
MF<-ModelFitStat(x,y,Beta) 84
 n1<-sum(y[y>0])
n1<-sum(y[y>0])85
 n<-nrow(y)
n<-nrow(y) 86
 LR<--2*(n1*log(n1)+(n-n1)*log(n-n1)-n*log(n))-MF$LOGL
LR<--2*(n1*log(n1)+(n-n1)*log(n-n1)-n*log(n))-MF$LOGL87
 LR_p_value<-pchisq(LR,df,lower.tail=FALSE)
LR_p_value<-pchisq(LR,df,lower.tail=FALSE)88
 LR_Test<-list(LR=LR,DF=df,LR_p_value=LR_p_value)
LR_Test<-list(LR=LR,DF=df,LR_p_value=LR_p_value)89

90
 ##compute Score
##compute Score91

92
 BetaIntercept<-matrix(c(as.vector(BetaIntercept),rep(0,ncol(x)-1)),ncol=1)
BetaIntercept<-matrix(c(as.vector(BetaIntercept),rep(0,ncol(x)-1)),ncol=1) 93
 obj<-funs(x,y,BetaIntercept)
obj<-funs(x,y,BetaIntercept) 94
 Score<-t(obj$U)%*%solve(obj$H)%*%obj$U
Score<-t(obj$U)%*%solve(obj$H)%*%obj$U95
 Score_p_value<-pchisq(Score,df,lower.tail=FALSE)
Score_p_value<-pchisq(Score,df,lower.tail=FALSE)96
 Score_Test<-list(Score=Score,DF=df,Score_p_value=Score_p_value)
Score_Test<-list(Score=Score,DF=df,Score_p_value=Score_p_value)97

98
 ##compute Wald test
##compute Wald test 99
 obj<-funs(x,y,Beta)
obj<-funs(x,y,Beta)100
 I_Diag<-diag((solve(obj$H)))
I_Diag<-diag((solve(obj$H))) 101

102
 Q<-matrix(c(rep(0,nrow(Beta)-1),as.vector(diag(rep(1,nrow(Beta)-1)))),nrow=nrow(Beta)-1)
Q<-matrix(c(rep(0,nrow(Beta)-1),as.vector(diag(rep(1,nrow(Beta)-1)))),nrow=nrow(Beta)-1)103
 Wald<-t(Q%*%Beta)%*%solve(Q%*%diag(I_Diag)%*%t(Q))%*%(Q%*%Beta)
Wald<-t(Q%*%Beta)%*%solve(Q%*%diag(I_Diag)%*%t(Q))%*%(Q%*%Beta) 104

105
 Wald_p_value<-pchisq(Wald,df,lower.tail=FALSE)
Wald_p_value<-pchisq(Wald,df,lower.tail=FALSE)106
 Wald_Test<-list(Wald=Wald,DF=df,Wald_p_value=Wald_p_value)
Wald_Test<-list(Wald=Wald,DF=df,Wald_p_value=Wald_p_value)107

108
 list(LR_Test=LR_Test,Score_Test=Score_Test,Wald_Test=Wald_Test)
list(LR_Test=LR_Test,Score_Test=Score_Test,Wald_Test=Wald_Test)109
 }
}110

111

 WhichEqual1<-function(x)
WhichEqual1<-function(x) {
{112
 a<-NULL
a<-NULL113

 for(i in 1:length(x))
for(i in 1:length(x)) {
{114

 if(x[i]==1)
if(x[i]==1) {
{115
 a<-c(a,i)
a<-c(a,i)116
 }
}117
 }
} 118
 a
a119
 }
} 120

121

 CheckOut<-function(source,check)
CheckOut<-function(source,check) {
{122

 for(j in 1:length(source))
for(j in 1:length(source)) {
{123

 for(k in 1:length(check))
for(k in 1:length(check)) {
{124
 if(source[j]==check[k])
if(source[j]==check[k])125
 source[j]<-0
source[j]<-0126
 }
} 127
 }
}128
 source[source>0]
source[source>0]129
 }
}130

131

 NegativeCheck<-function(x)
NegativeCheck<-function(x) {
{132

 for(i in length(x):1)
for(i in length(x):1) {
{133
 if(x[i]>0)
if(x[i]>0)134
 break
break135
 }
}136
 i
i137
 }
}138

139

 CycleCheck<-function(x)
CycleCheck<-function(x) {
{140
 NegativeFlg<-NegativeCheck(x)
NegativeFlg<-NegativeCheck(x)141

 if(NegativeFlg==length(x))
if(NegativeFlg==length(x)) {
{142
 return(FALSE)
return(FALSE)143
 }
} 144
 NegVec<-x[(NegativeFlg+1):length(x)]
NegVec<-x[(NegativeFlg+1):length(x)]145
 PosVec<-x[(2*NegativeFlg-length(x)+1):NegativeFlg]
PosVec<-x[(2*NegativeFlg-length(x)+1):NegativeFlg]146

147
 NegVec<-sort(-1*NegVec)
NegVec<-sort(-1*NegVec)148
 PosVec<-sort(PosVec)
PosVec<-sort(PosVec)149

150
 if(all((NegVec-PosVec)==0))
if(all((NegVec-PosVec)==0))151
 return(TRUE)
return(TRUE)152

153
 return(FALSE)
return(FALSE) 154
 }
} 155

156

157
 ##stepwise
##stepwise158

 Stepwise<-function(x,y,checkin_pvalue=0.05,checkout_pvalue=0.05)
Stepwise<-function(x,y,checkin_pvalue=0.05,checkout_pvalue=0.05) {
{159
 ##as matrix
##as matrix160
 x<-matrix(as.vector(x),nrow=nrow(y))
x<-matrix(as.vector(x),nrow=nrow(y))161
 ##indication of variable
##indication of variable162
 indict<-rep(0,ncol(x)) ##which column enter
indict<-rep(0,ncol(x)) ##which column enter163
 ##intercept entered
##intercept entered164
 indict[1]<-1
indict[1]<-1165
 Beta<-NULL
Beta<-NULL166
 print("Intercept Entered
print("Intercept Entered ")
")167
 Result<-Newtons(funs,x[,1],y)
Result<-Newtons(funs,x[,1],y)168
 Beta<-Result$Beta
Beta<-Result$Beta169
 BetaIntercept<-Result$Beta
BetaIntercept<-Result$Beta 170
 uni_matrix<-matrix(rep(1,nrow(y)),nrow= nrow(y));
uni_matrix<-matrix(rep(1,nrow(y)),nrow= nrow(y));171
 LOGL<--2*(t(y)%*%log(pi_fun(x[,1],Beta))+t(uni_matrix-y)%*%log(uni_matrix-pi_fun(x[,1],Beta)))
LOGL<--2*(t(y)%*%log(pi_fun(x[,1],Beta))+t(uni_matrix-y)%*%log(uni_matrix-pi_fun(x[,1],Beta)))172
 print(paste("-2Log=",LOGL))
print(paste("-2Log=",LOGL)) 173
 indexVector<-WhichEqual1(indict)
indexVector<-WhichEqual1(indict) 174
 ##check other variable
##check other variable175

176
 VariableFlg<-NULL
VariableFlg<-NULL177
 Terminate<-FALSE
Terminate<-FALSE178

 repeat
repeat {
{ 179

 if(Terminate==TRUE)
if(Terminate==TRUE) {
{180
 print("Model building terminates because the last effect entered is removed by the Wald statistic criterion. ")
print("Model building terminates because the last effect entered is removed by the Wald statistic criterion. ")181
 break
break182
 }
}183

184
 pvalue<-rep(1,ncol(x))
pvalue<-rep(1,ncol(x))185

186
 k<-2:length(indict)
k<-2:length(indict)187
 k<-CheckOut(k,indexVector)
k<-CheckOut(k,indexVector) 188

 for(i in k)
for(i in k) {
{189

190
 obj<-funs(c(x[,indexVector],x[,i]),y,c(as.vector(Beta),0))
obj<-funs(c(x[,indexVector],x[,i]),y,c(as.vector(Beta),0)) 191
 Score<-t(obj$U)%*%solve(obj$H,obj$U)
Score<-t(obj$U)%*%solve(obj$H,obj$U) 192
 Score_pvalue<-pchisq(Score,1,lower.tail=FALSE)
Score_pvalue<-pchisq(Score,1,lower.tail=FALSE) 193
 pvalue[i]<-Score_pvalue
pvalue[i]<-Score_pvalue 194
 }
}195
 #print("Score pvalue for variable enter")
#print("Score pvalue for variable enter")196
 #print(pvalue)
#print(pvalue)197
 pvalue_min<-min(pvalue)
pvalue_min<-min(pvalue)198

 if(pvalue_min<checkin_pvalue)
if(pvalue_min<checkin_pvalue) {
{199
 j<-which.min(pvalue)
j<-which.min(pvalue) 200

201
 print(paste(x_name[j]," entered:"))
print(paste(x_name[j]," entered:"))202
 ##set indication of variable
##set indication of variable203
 indict[j]<-1
indict[j]<-1204
 VariableFlg<-c(VariableFlg,j)
VariableFlg<-c(VariableFlg,j) 205

206
 indexVector<-WhichEqual1(indict)
indexVector<-WhichEqual1(indict)207
 print("indexVector--test")
print("indexVector--test")208
 print(indexVector)
print(indexVector) 209

210
 Result<-Newtons(funs,x[,indexVector],y)
Result<-Newtons(funs,x[,indexVector],y)211
 Beta<-NULL
Beta<-NULL212
 Beta<-Result$Beta
Beta<-Result$Beta 213

214
 ##compute model fit statistics
##compute model fit statistics215
 print("Model Fit Statistics")
print("Model Fit Statistics")216
 MFStat<-ModelFitStat(x[,indexVector],y,Beta)
MFStat<-ModelFitStat(x[,indexVector],y,Beta)217
 print(MFStat)
print(MFStat)218
 ##test globel null hypothesis:Beta=0
##test globel null hypothesis:Beta=0 219
 print("Testing Global Null Hypothese:Beta=0")
print("Testing Global Null Hypothese:Beta=0") 220
 GNTest<-GlobalNullTest(x[,indexVector],y,Beta,BetaIntercept)
GNTest<-GlobalNullTest(x[,indexVector],y,Beta,BetaIntercept)221
 print(GNTest)
print(GNTest) 222

223

 repeat
repeat {
{224
 ##compute Wald test in order to remove variable
##compute Wald test in order to remove variable225
 indexVector<-WhichEqual1(indict)
indexVector<-WhichEqual1(indict)226
 obj<-funs(x[,indexVector],y,Beta)
obj<-funs(x[,indexVector],y,Beta)227
 H_Diag<-sqrt(diag(solve(obj$H)))
H_Diag<-sqrt(diag(solve(obj$H))) 228
 WaldChisq<-(as.vector(Beta)/H_Diag)^2
WaldChisq<-(as.vector(Beta)/H_Diag)^2229
 WaldChisqPvalue<-pchisq(WaldChisq,1,lower.tail=FALSE)
WaldChisqPvalue<-pchisq(WaldChisq,1,lower.tail=FALSE)230
 WaldChisqTest<-list(WaldChisq=WaldChisq,WaldChisqPvalue=WaldChisqPvalue)
WaldChisqTest<-list(WaldChisq=WaldChisq,WaldChisqPvalue=WaldChisqPvalue)231
 print("Wald chisq pvalue for variable remove")
print("Wald chisq pvalue for variable remove")232
 print(WaldChisqPvalue)
print(WaldChisqPvalue)233

234
 ##check wald to decide to which variable to be removed
##check wald to decide to which variable to be removed235
 pvalue_max<-max(WaldChisqPvalue[2:length(WaldChisqPvalue)])##not check for intercept
pvalue_max<-max(WaldChisqPvalue[2:length(WaldChisqPvalue)])##not check for intercept236

237

 if(pvalue_max>checkout_pvalue)
if(pvalue_max>checkout_pvalue) {
{238
 n<-which.max(WaldChisqPvalue[2:length(WaldChisqPvalue)])##not check for intercept
n<-which.max(WaldChisqPvalue[2:length(WaldChisqPvalue)])##not check for intercept239
 print(paste(x_name[indexVector[n+1]]," is removed:"))
print(paste(x_name[indexVector[n+1]]," is removed:"))240
 ##set indication of variable
##set indication of variable241

242
 indict[indexVector[n+1]]<-0
indict[indexVector[n+1]]<-0 243
 m<- indexVector[n+1]
m<- indexVector[n+1] 244
 VariableFlg<-c(VariableFlg,-m)
VariableFlg<-c(VariableFlg,-m) 245

246
 ##renew Beta
##renew Beta247
 indexVector<-WhichEqual1(indict)
indexVector<-WhichEqual1(indict)248
 Result<-Newtons(funs,x[,indexVector],y)
Result<-Newtons(funs,x[,indexVector],y)249
 Beta<-NULL
Beta<-NULL250
 Beta<-Result$Beta
Beta<-Result$Beta 251

252

 if(CycleCheck(VariableFlg)==TRUE)
if(CycleCheck(VariableFlg)==TRUE) {
{253
 Terminate<-TRUE
Terminate<-TRUE254
 break
break255
 }
} 256

257
 }
}258

 else
else  {
{259
 print(paste("No (additional) effects met the" ,checkout_pvalue,"significance level for removal from the model."))
print(paste("No (additional) effects met the" ,checkout_pvalue,"significance level for removal from the model."))260
 break;
break;261
 }
} 262
 }##repeat end
}##repeat end263
 }
}264

 else
else  {
{265
 print(paste("No effects met the" ,checkin_pvalue," significance level for entry into the model"))
print(paste("No effects met the" ,checkin_pvalue," significance level for entry into the model"))266
 break
break267
 }
}268
 }##repeat end
}##repeat end 269

270
 ##
##271
 print("Beta")
print("Beta")272
 print(Beta)
print(Beta)273
 print("Analysis of Maximum Likelihood Estimates")
print("Analysis of Maximum Likelihood Estimates") 274
 obj<-funs(x[,indexVector],y,Beta)
obj<-funs(x[,indexVector],y,Beta)275
 H_Diag<-sqrt(diag(solve(obj$H)))
H_Diag<-sqrt(diag(solve(obj$H))) 276
 WaldChisq<-(as.vector(Beta)/H_Diag)^2
WaldChisq<-(as.vector(Beta)/H_Diag)^2277
 WaldChisqPvalue<-pchisq(WaldChisq,1,lower.tail=FALSE)
WaldChisqPvalue<-pchisq(WaldChisq,1,lower.tail=FALSE)278
 WaldChisqTest<-list(WaldChisq=WaldChisq,WaldChisqPvalue=WaldChisqPvalue)
WaldChisqTest<-list(WaldChisq=WaldChisq,WaldChisqPvalue=WaldChisqPvalue) 279
 print(WaldChisqTest)
print(WaldChisqTest)280
 }
} 使用例子
1
 #example5--stepwise
#example5--stepwise2
 rs<-read.csv("diabetes.csv",head=TRUE)
rs<-read.csv("diabetes.csv",head=TRUE) 3
 x<-c(rep(1,768),rs[,1],rs[,2],rs[,3],rs[,4],rs[,5],rs[,6],rs[,7],rs[,8])
x<-c(rep(1,768),rs[,1],rs[,2],rs[,3],rs[,4],rs[,5],rs[,6],rs[,7],rs[,8]) 4
 x<-matrix(x,nrow=768)
x<-matrix(x,nrow=768)5
 y<-matrix(c(rs[,9]),nrow=768)
y<-matrix(c(rs[,9]),nrow=768)6
 x_name<-c("INTERCEPT","PREGNANT","PLASMA","PRESSURE","TRICEPS","INSULIN","INDEX","PEDIGREE","AGE")
x_name<-c("INTERCEPT","PREGNANT","PLASMA","PRESSURE","TRICEPS","INSULIN","INDEX","PEDIGREE","AGE")7
 Stepwise(x,y)
Stepwise(x,y)



