这涉及一系列文献:
文献1: Locality and Hard SAT-Instances
——Markström, Klas. ‘Locality and Hard SAT-Instances’. Journal on Satisfiability, Boolean Modeling and Computation, vol. 2, no. 1-4, pp. 221-227, 2006
求解器如何尝试避免这些实例所带来的至少一些陷阱。
AbstractIn this note we construct a family of SAT-instance based on Eulerian graphs which are aimed at being hard for resolution based SAT-solvers. We discuss some experiments made with instances of this type and how a solver can try to avoid at least some of the pitfalls presented by these instances. Finally we look at how the density of subformulae can influence the hardness of SAT instances. 我们将讨论使用这种类型的实例所做的一些实验,以及求解器如何尝试避免这些实例所带来的至少一些陷阱。 |
|
|
1. Introduction DPLL method 这个方法实际上执行深度优先搜索,并结合单元子句的传播。 该方法是一个多项式时间复杂度,等价于树状归结。 Tree-like resolution is a fairly weak proof system and for a long time the performance of solvers were quite limited..树形归结是一种较弱的证明系统,长期以来求解器的性能都很有限。
树状归结弱于常规归结(包括many proper refinements of general resolution)和戴维斯-普特南归结。
The aim of this note is to construct a family of CNF formulae which are specially tuned to make use of the structure of current solvers to produce small hard examples. 这篇笔记的目的是构造一组CNF公式,这些公式是特别调整的,以利用当前求解器的结构来产生小的硬例子。 Several instances from the constructed family were submitted as benchmarks for the 2005 SAT solver competition, none of which could be solved by the competing solvers. 构建族中的几个实例被提交为2005年SAT求解器竞赛的基准,没有一个可以被竞争求解器解决。 In connection with this we will also give a short discussion of the importance of local dense subformulae for the performance of resolution based solvers. 与此相关,我们还将简要讨论局部密集子公式对基于分辨率的求解器性能的重要性。 Here we will also comment on some structural similarities between our family of formulae and formulae from the random k-SAT distribution. 在这里,我们还将评论我们的公式家族和随机k-SAT分布的公式之间的一些结构相似性。 |
|
|
2. The problem class 图着色问题及其形式化方法
对应生成cnf的形式化方法实现 coloring.py 1 #!/usr/bin/env python 2 # -*- coding:utf-8 -*- 3 """Formulas that encode coloring related problems 4 """ 5 6 from cnfgen.formula.cnf import CNF 7 from cnfgen.graphs import Graph 8 from cnfgen.localtypes import non_negative_int 9 10 def GraphColoringFormula(G, colors, functional=True, formula_class=CNF): 11 """Generates the clauses for colorability formula 12 13 The formula encodes the fact that the graph :math:`G` has a coloring 14 with color set ``colors``. This means that it is possible to 15 assign one among the elements in ``colors``to that each vertex of 16 the graph such that no two adjacent vertices get the same color. 17 18 Parameters 19 ---------- 20 G : cnfgen.Graph 21 a simple undirected graph 22 colors : non negative int 23 the number of colors 24 functional: bool 25 forbid a vertex to be mapped to multiple colors 26 27 Returns 28 ------- 29 CNF 30 the CNF encoding of the coloring problem on graph ``G`` 31 32 """ 33 non_negative_int(colors, 'colors') 34 G = Graph.normalize(G, 'G') 35 36 # Describe the formula 37 description = "Graph {}-Colorability of {}".format(colors,G) 38 F = formula_class(description=description) 39 col = F.new_mapping(G.order(), colors,label='x_{{{0}{1}}}') 40 41 # Color each vertex 42 F.force_complete_mapping(col) 43 if functional: 44 F.force_functional_mapping(col) 45 46 47 # This is a legal coloring 48 for (v1, v2) in G.edges(): 49 for c in range(1,colors+1): 50 F.add_clause([-col(v1, c), -col(v2, c)]) 51 52 return F 53 54 55 def EvenColoringFormula(G, formula_class=CNF): 56 """Even coloring formula 57 58 The formula is defined on a graph :math:`G` and claims that it is 59 possible to split the edges of the graph in two parts, so that 60 each vertex has an equal number of incident edges in each part. 61 62 The formula is defined on graphs where all vertices have even 63 degree. The formula is satisfiable only on those graphs with an 64 even number of edges in each connected component [1]_. 65 66 Arguments 67 --------- 68 G : cnfgen.Graph 69 a simple undirected graph where all vertices have even degree 70 71 Raises 72 ------ 73 ValueError 74 if the graph in input has a vertex with odd degree 75 76 Returns 77 ------- 78 CNF object 79 80 References 81 ---------- 82 .. [1] Locality and Hard SAT-instances, Klas Markstrom 83 Journal on Satisfiability, Boolean Modeling and Computation 2 (2006) 221-228 84 85 """ 86 G = Graph.normalize(G, 'G') 87 88 description = "Even coloring formula on " + G.name 89 F = formula_class(description=description) 90 91 e = F.new_graph_edges(G) 92 93 # Defined on both side 94 for v in G.vertices(): 95 96 if G.degree(v) % 2 == 1: 97 raise ValueError( 98 "Markstrom's Even Coloring formulas requires all\n
cnf.py 1 #!/usr/bin/env python 2 # -*- coding:utf-8 -*- 3 """Build and manipulate CNF formulas 4 5 The module `contains facilities to generate cnf formulas, in order to 6 be printed in DIMACS, OPB or LaTeX formats. Such formulas are ready to 7 be fed to sat solvers. 8 9 The module implements the `CNF` object, which is the main entry point 10 to the `cnfgen` library. 11 12 Copyright (C) 2012-2022 Massimo Lauria <lauria.massimo@gmail.com> 13 https://github.com/MassimoLauria/cnfgen.git 14 15 """ 16 from cnfgen.formula.cnfio import CNFio 17 from cnfgen.formula.linear import CNFLinear 18 from cnfgen.formula.variables import VariablesManager 19 20 21 class CNF(VariablesManager, CNFio, CNFLinear): 22 """Propositional formulas in conjunctive normal form. 23 24 A CNF formula is a sequence of clauses, which are sequences of 25 literals. Each literal is either a variable or its negation. 26 27 Use ``add_clause`` to add new clauses to CNF. Clauses will be added 28 multiple times in case of multiple insertion of the same clauses. 29 30 For documentation purpose it is possible use have an additional 31 comment header at the top of the formula, which will be 32 *optionally* exported to LaTeX or dimacs. 33 34 Implementation: for efficiency reason clauses and variable can 35 only be added, and not deleted. Furthermore order matters in 36 the representation. 37 38 Examples 39 -------- 40 >>> c=CNF([[1, 2, -3], [-2, 4]]) 41 >>> print( c.to_dimacs(),end='') 42 p cnf 4 2 43 1 2 -3 0 44 -2 4 0 45 >>> c.add_clause([-3, 4, -5]) 46 >>> print( c.to_dimacs(),end='') 47 p cnf 5 3 48 1 2 -3 0 49 -2 4 0 50 -3 4 -5 0 51 >>> print(c[1]) 52 [-2, 4] 53 """ 54 55 def __init__(self, clauses=None, description=None): 56 """Propositional formulas in conjunctive normal form. 57 58 Parameters 59 ---------- 60 clauses : ordered list of clauses 61 a clause with k literals list containing k pairs, each 62 representing a literal (see `add_clause`). First element 63 is the polarity and the second is the variable, which must 64 be an hashable object. 65 66 E.g. (not x3) or x4 or (not x2) is encoded

1 #!/usr/bin/env python 2 # -*- coding:utf-8 -*- 3 """Utilities to manage graph formats and graph files in order to build 4 formulas that are graph based. 5 """ 6 7 import os 8 import io 9 import random 10 from io import StringIO 11 import copy 12 from bisect import bisect_right, bisect_left 13 14 import networkx 15 16 from cnfgen.localtypes import positive_int, non_negative_int 17 18 __all__ = [ 19 "readGraph", "writeGraph", 20 "Graph", "DirectedGraph", "BipartiteGraph", 21 "supported_graph_formats", 22 "bipartite_random_left_regular", "bipartite_random_regular", 23 "bipartite_random_m_edges", "bipartite_random", 24 "dag_complete_binary_tree", "dag_pyramid", "dag_path" 25 ] 26 27 ################################################################# 28 # Import third party code 29 ################################################################# 30 31 32 class BipartiteEdgeList(): 33 """Edge list for bipartite graphs""" 34 35 def __init__(self, B): 36 self.B = B 37 38 def __len__(self): 39 return self.B.number_of_edges() 40 41 def __contains__(self, t): 42 return len(t) == 2 and self.B.has_edge(t[0], t[1]) 43 44 def __iter__(self): 45 for u in range(1, self.B.left_order() + 1): 46 yield from ((u, v) for v in self.B.right_neighbors(u)) 47 48 49 class GraphEdgeList(): 50 """Edge list for bipartite graphs""" 51 52 def __init__(self, G): 53 self.G = G 54 55 def __len__(self): 56 return self.G.number_of_edges() 57 58 def __contains__(self, t): 59 return len(t) == 2 and self.G.has_edge(t[0], t[1]) 60 61 def __iter__(self): 62 n = self.G.number_of_vertices() 63 G = self.G 64 for u in range(1, n): 65 pos = bisect_right(G.adjlist[u], u) 66 while pos < len(G.adjlist[u]): 67 v = G.adjlist[u][pos] 68 yield (u, v) 69 pos += 1 70 71 72 class DirectedEdgeList(): 73 """Edge list for bipartite graphs""" 74 75 def __init__(self, D, sort_by_predecessors=True): 76 self.D = D 77 self.sort_by_pred = sort_by_predecessors 78 79 def __len__(self): 80 return self.D.number_of_edges() 81 82 def __contains__(self, t): 83 return len(t) == 2 and self.D.has_edge(t[0], t[1]) 84 85 def __iter__(self): 86 n = self.D.number_of_vertices() 87 if self.sort_by_pred: 88 successors = self.D.succ 89 for src in range(1, n+1): 90 for dest in successors[src]: 91 yield (src, dest) 92 else: 93 predecessors = self.D.pred 94 for dest in range(1, n+1): 95 for src in predecessors[dest]: 96 yield (src, dest) 97 98 99 class BaseGraph(): 100 """Base class for graphs""" 101 102 def is_dag(self): 103 """Test whether the graph is directed acyclic 104 105 This is not a full test. It only checks that all directed edges (u,v) 106 have that u < v.""" 107 raise NotImplementedError 108 109 def is_directed(self): 110 "Test whether the graph is directed" 111 raise NotImplementedError 112 113 def is_multigraph(self): 114 "Test whether the graph can have multi-edges" 115 return False 116 117 def is_bipartite(self): 118 "Test whether the graph is a bipartite object" 119 return False 120 121 def order(self): 122 return self.number_of_vertices() 123 124 def vertices(self): 125 return range(1, self.number_of_vertices()+1) 126 127 def number_of_vertices(self): 128 raise NotImplementedError 129 130 def number_of_edges(self): 131 raise NotImplementedError 132 133 def has_edge(self, u, v): 134 raise NotImplementedError 135 136 def add_edge(self, u, v): 137 raise NotImplementedError 138 139 def add_edges_from(self, edges): 140 for u, v in edges: 141 self.add_edge(u, v) 142 143 def edges(self): 144 raise NotImplementedError 145 146 def __len__(self): 147 return self.number_of_vertices() 148 149 def to_networkx(self): 150 """Convert the graph TO a networkx object.""" 151 raise NotImplementedError 152 153 @classmethod 154 def from_networkx(cls, G): 155 """Create a graph object from a networkx graph""" 156 raise NotImplementedError 157 158 @classmethod 159 def normalize(cls, G): 160 """Guarantees a cnfgen graph object""" 161 raise NotImplementedError 162 163 @classmethod 164 def supported_file_formats(cls): 165 """File formats supported for graph I/O""" 166 raise NotImplementedError 167 168 @classmethod 169 def graph_type_name(cls): 170 """File formats supported for graph I/O""" 171 raise NotImplementedError 172 173 @classmethod 174 def from_file(cls, fileorname, fileformat=None): 175 """Load the graph from a file 176 177 The file format is either indicated in the `fileformat` variable or, if 178 that is `None`, or from the extension of the filename. 179 180 Parameters 181 ----------- 182 fileorname: str or file-like object 183 the input file from which the graph is read. If it is a string 184 then the graph is read from a file with that string as 185 filename. Otherwise if the fileorname is a file object (or 186 a text stream), the graph is read from there. 187 188 Input files are assumed to be UTF-8 by default (for some 189 formats it is actually ascii) 190 191 fileformat: string, optional 192 The file format that the parser should expect to receive. 193 See also :py:func:`cnfgen.graph.supported_formats`. By default 194 it tries to autodetect it from the file name extension (when applicable).""" 195 196 # Reduce to the case of filestream 197 if isinstance(fileorname, str): 198 with open(fileorname, 'r', encoding='utf-8') as file_handle: 199 return cls.from_file(file_handle, fileformat) 200 201 # Discover and test file format 202 fileformat = guess_fileformat(fileorname, fileformat) 203 allowed = cls.supported_file_formats() 204 typename = cls.graph_type_name() 205 if fileformat not in allowed: 206 raise ValueError( 207 "Invalid file type." 208 " For {} graphs we support {}".format(typename, 209 allowed)) 210 211 # Read file 212 return readGraph(fileorname, typename, fileformat) 213 214 215 class Graph(BaseGraph): 216 217 def is_dag(self): 218 return False 219 220 def is_directed(self): 221 return False 222 223 def __init__(self, n, name=None): 224 non_negative_int(n, 'n') 225 self.n = n 226 self.m = 0 227 self.adjlist = [[] for i in range(n+1)] 228 self.edgeset = set() 229 if name is None: 230 self.name = "a simple graph with {} vertices".format(n) 231 else: 232 self.name = name 233 234 def add_edge(self, u, v): 235 if not (1 <= u <= self.n and 1 <= v <= self.n and u != v): 236 raise ValueError( 237 "u,v must be distinct, between 1 and the number of nodes") 238 if (u, v) in self.edgeset: 239 return 240 u, v = min(u, v), max(u, v) 241 pos = bisect_right(self.adjlist[u], v) 242 self.adjlist[u].insert(pos, v) 243 pos = bisect_right(self.adjlist[v], u) 244 self.adjlist[v].insert(pos, u) 245 self.m += 1 246 self.edgeset.add((u, v)) 247 self.edgeset.add((v, u)) 248 249 def update_vertex_number(self, new_value): 250 """Raises the number of vertices to `new_value`""" 251 non_negative_int(new_value, 'new_value') 252 for _ in range(self.n,new_value): 253 self.adjlist.append([]) 254 self.n = max(self.n, new_value) 255 256 def remove_edge(self,u,v): 257 if not self.has_edge(u,v): 258 return 259 self.edgeset.remove((u,v)) 260 self.edgeset.remove((v,u)) 261 self.adjlist[u].remove(v) 262 self.adjlist[v].remove(u) 263 self.m -= 1 264 265 def has_edge(self, u, v): 266 return (u, v) in self.edgeset 267 268 def vertices(self): 269 return range(1, self.n+1) 270 271 def edges(self): 272 """Outputs all edges in the graph""" 273 return GraphEdgeList(self) 274 275 def number_of_vertices(self): 276 return self.n 277 278 def number_of_edges(self): 279 return self.m 280 281 def to_networkx(self): 282 G = networkx.Graph() 283 G.add_nodes_from(range(1, self.n+1)) 284 G.add_edges_from(self.edges()) 285 return G 286 287 def neighbors(self, u): 288 """Outputs the neighbors of vertex `u` 289 290 The sequence of neighbors is guaranteed to be sorted. 291 """ 292 if not(1 <= u <= self.n): 293 raise ValueError("vertex u not in the graph") 294 yield from self.adjlist[u] 295 296 def degree(self, u): 297 if not(1 <= u <= self.n): 298 raise ValueError("vertex u not in the graph") 299 return len(self.adjlist[u]) 300 301 @classmethod 302 def from_networkx(cls, G): 303 if not isinstance(G, networkx.Graph): 304 raise ValueError('G is expected to be of type networkx.Graph') 305 G = normalize_networkx_labels(G) 306 C = cls(G.order()) 307 C.add_edges_from(G.edges()) 308 try: 309 C.name = G.name 310 except AttributeError: 311 C.name = '<unknown graph>' 312 return C 313 314 @classmethod 315 def graph_type_name(cls): 316 """Simple graphs are laleled as 'simple'""" 317 return 'simple' 318 319 @classmethod 320 def supported_file_formats(cls): 321 """File formats supported for simple graph I/O""" 322 # Check that DOT is a supported format 323 if has_dot_library(): 324 return ['kthlist', 'gml', 'dot', 'dimacs'] 325 else: 326 return ['kthlist', 'gml', 'dimacs'] 327 328 @classmethod 329 def null_graph(cls): 330 return cls(0, 'the null graph') 331 332 @classmethod 333 def empty_graph(cls, n): 334 return cls(n, 'the empty graph of order '+str(n)) 335 336 @classmethod 337 def complete_graph(cls, n): 338 G = cls(n, 'the complete graph of order '+str(n)) 339 for u in range(1, n): 340 for v in range(u+1, n+1): 341 G.add_edge(u, v) 342 return G 343 344 @classmethod 345 def star_graph(cls, n): 346 G = cls(n+1, 'the star graph with {} arms'.format(n)) 347 for u in range(1, n+1): 348 G.add_edge(u, n+1) 349 return G 350 351 @classmethod 352 def normalize(cls, G, varname=''): 353 """Guarantees a cnfgen.graphs.Graph object 354 355 If the given graph `G` is a networkx.Graph object, this method 356 produces a CNFgen simple graph object, relabeling vertices so that 357 vertices are labeled as numbers from 1 to `n`, where `n` is the number 358 of vertices in `G`. If the vertices in the original graph have some 359 kind of order, the order is preserved. 360 361 If `G` is already a `cnfgen.graphs.Graph` object, nothing is done. 362 363 Parameters 364 ---------- 365 cls: a class 366 367 G : networkx.Graph or cnfgen.Graph 368 the graph to normalize/check 369 varname: str 370 the variable name, for error messages (default: 'G') 371 """ 372 typemsg = "type of argument '{}' must be either networx.Graph or cnfgen.Graph" 373 conversionmsg = "cannot convert '{}' into a cnfgen.Graph object" 374 if not isinstance(G, (Graph, networkx.Graph)): 375 raise TypeError(typemsg.format(varname)) 376 if isinstance(G, Graph): 377 return G 378 try: 379 G2 = cls.from_networkx(G) 380 return G2 381 except AttributeError: 382 raise ValueError(conversionmsg.format(varname)) 383 384 385 class DirectedGraph(BaseGraph): 386 387 def is_dag(self): 388 """Is the graph acyclic? 389 390 The vertices in the graph are assumed to be topologically sorted, 391 therefore this function just determines whether there are edges going 392 backward with respect to this order, which can be done in O(1) because 393 edges can be added and not removed.""" 394 return self.still_a_dag 395 396 def is_directed(self): 397 return True 398 399 def __init__(self, n, name='a simple directed graph'): 400 non_negative_int(n, 'n') 401 self.n = n 402 self.m = 0 403 self.edgeset = set() 404 self.still_a_dag = True 405 self.pred = [[] for i in range(n+1)] 406 self.succ = [[] for i in range(n+1)] 407 if name is None: 408 self.name = "a directed graph with {} vertices".format(n) 409 else: 410 self.name = name 411 412 def add_edge(self, src, dest): 413 if not (1 <= src <= self.n and 1 <= dest <= self.n): 414 raise ValueError( 415 "u,v must be distinct, between 1 and the number of nodes") 416 if self.has_edge(src, dest): 417 return 418 if src >= dest: 419 self.still_a_dag = False 420 421 pos = bisect_right(self.pred[dest], src) 422 self.pred[dest].insert(pos, src) 423 424 pos = bisect_right(self.succ[src], dest) 425 self.succ[src].insert(pos, dest) 426 427 self.m += 1 428 self.edgeset.add((src, dest)) 429 430 def has_edge(self, src, dest): 431 """True if graph contains directed edge (src,dest)""" 432 return (src, dest) in self.edgeset 433 434 def vertices(self): 435 return range(1, self.n+1) 436 437 def edges(self): 438 return DirectedEdgeList(self) 439 440 def edges_ordered_by_successors(self): 441 return DirectedEdgeList(self, sort_by_predecessors=False) 442 443 def number_of_vertices(self): 444 return self.n 445 446 def number_of_edges(self): 447 return self.m 448 449 def to_networkx(self): 450 G = networkx.DiGraph() 451 G.add_nodes_from(range(1, self.n+1)) 452 G.add_edges_from(self.edges()) 453 return G 454 455 def predecessors(self, u): 456 """Outputs the predecessors of vertex `u` 457 458 The sequence of predecessors is guaranteed to be sorted.""" 459 if not(1 <= u <= self.n): 460 raise ValueError("vertex u not in the graph") 461 yield from self.pred[u] 462 463 def successors(self, u): 464 """Outputs the successors of vertex `u` 465 466 The sequence of successors is guaranteed to be sorted.""" 467 if not(1 <= u <= self.n): 468 raise ValueError("vertex u not in the graph") 469 yield from self.succ[u] 470 471 def in_degree(self, u): 472 if not(1 <= u <= self.n): 473 raise ValueError("vertex u not in the graph") 474 return len(self.pred[u]) 475 476 def out_degree(self, v): 477 if not(1 <= v <= self.n): 478 raise ValueError("vertex v not in the graph") 479 return len(self.succ[v]) 480 481 @classmethod 482 def from_networkx(cls, G): 483 if not isinstance(G, networkx.DiGraph): 484 raise ValueError('G is expected to be of type networkx.DiGraph') 485 G = normalize_networkx_labels(G) 486 C = cls(G.order()) 487 C.add_edges_from(G.edges()) 488 try: 489 C.name = G.name 490 except AttributeError: 491 C.name = '<unknown graph>' 492 return C 493 494 @classmethod 495 def graph_type_name(cls): 496 """Directed graphs are laleled as 'digraph'""" 497 return 'digraph' 498 499 @classmethod 500 def supported_file_formats(cls): 501 """File formats supported for directed graph I/O""" 502 if has_dot_library(): 503 return ['kthlist', 'gml', 'dot', 'dimacs'] 504 else: 505 return ['kthlist', 'gml', 'dimacs'] 506 507 @classmethod 508 def normalize(cls, G, varname='G'): 509 """Guarantees a cnfgen.graphs.DirerctedGraph object 510 511 If the given graph `G` is a networkx.DiGraph object, this method 512 produces a CNFgen directed graph object, relabeling vertices so that 513 vertices are labeled as numbers from 1 to `n`, where `n` is the number 514 of vertices in `G`. If the vertices in the original graph have some 515 kind of order, the order is preserved. 516 517 If all edges go from lower vertices to higher vertices, with respect 518 to the labeling, then t he graph is considered a directed acyclic 519 graph DAG. 520 521 If `G` is already a `cnfgen.graphs.DirectedGraph` object, nothing is done. 522 523 Parameters 524 ---------- 525 cls: a class 526 527 G : networkx.DiGraph or cnfgen.DirectedGraph 528 the graph to normalize/check 529 varname: str 530 the variable name, for error messages (default: 'G') 531 """ 532 typemsg = "type of argument '{}' must be either networx.DiGraph or cnfgen.DirectedGraph" 533 conversionmsg = "cannot convert '{}' into a cnfgen.DirectedGraph object" 534 if not isinstance(G, (DirectedGraph, networkx.DiGraph)): 535 raise TypeError(typemsg.format(varname)) 536 if isinstance(G, DirectedGraph): 537 return G 538 try: 539 G2 = cls.from_networkx(G) 540 return G2 541 except AttributeError: 542 raise ValueError(conversionmsg.format(varname)) 543 544 545 class BaseBipartiteGraph(BaseGraph): 546 """Base class for bipartite graphs""" 547 548 def __init__(self, L, R, name=None): 549 non_negative_int(L, 'L') 550 non_negative_int(R, 'R') 551 self.lorder = L 552 self.rorder = R 553 if name is None: 554 self.name = 'a bipartite graph with ({},{}) vertices'.format(L, R) 555 else: 556 self.name = name 557 558 def is_bipartite(self): 559 return True 560 561 def number_of_vertices(self): 562 return self.lorder + self.rorder 563 564 def edges(self): 565 return BipartiteEdgeList(self) 566 567 def left_order(self): 568 return self.lorder 569 570 def right_order(self): 571 return self.rorder 572 573 def left_degree(self, v): 574 return len(self.left_neighbors(v)) 575 576 def right_degree(self, u): 577 return len(self.right_neighbors(u)) 578 579 def left_neighbors(self, v): 580 raise NotImplementedError 581 582 def right_neighbors(self, u): 583 raise NotImplementedError 584 585 def parts(self): 586 return range(1, self.lorder + 1), range(1, self.rorder + 1) 587 588 def to_networkx(self): 589 G = networkx.Graph() 590 n, m = self.lorder, self.rorder 591 G.add_nodes_from(range(1, n+1), bipartite=0) 592 G.add_nodes_from(range(n+1, m+n+1), bipartite=1) 593 G.add_edges_from((u, v+n) for (u, v) in self.edges()) 594 G.name = self.name 595 return G 596 597 598 class BipartiteGraph(BaseBipartiteGraph): 599 def __init__(self, L, R, name=None): 600 non_negative_int(L, 'L') 601 non_negative_int(R, 'R') 602 BaseBipartiteGraph.__init__(self, L, R, name) 603 self.ladj = {} 604 self.radj = {} 605 self.edgeset = set() 606 607 def has_edge(self, u, v): 608 return (u, v) in self.edgeset 609 610 def add_edge(self, u, v): 611 """Add an edge to the graph. 612 613 - multi-edges are not allowed 614 - neighbors of a vertex are kept in numberic order 615 616 Examples 617 -------- 618 >>> G = BipartiteGraph(3,5) 619 >>> G.add_edge(2,3) 620 >>> G.add_edge(2,2) 621 >>> G.add_edge(2,3) 622 >>> G.right_neighbors(2) 623 [2, 3] 624 """ 625 if not (1 <= u <= self.lorder and 1 <= v <= self.rorder): 626 raise ValueError("Invalid choice of vertices") 627 628 if (u, v) in self.edgeset: 629 return 630 631 if u not in self.ladj: 632 self.ladj[u] = [] 633 if v not in self.radj: 634 self.radj[v] = [] 635 636 pv = bisect_right(self.ladj[u], v) 637 pu = bisect_right(self.radj[v], u) 638 self.ladj[u].insert(pv, v) 639 self.radj[v].insert(pu, u) 640 self.edgeset.add((u, v)) 641 642 def number_of_edges(self): 643 return len(self.edgeset) 644 645 def right_neighbors(self, u): 646 """Outputs the neighbors of a left vertex `u` 647 648 The sequence of neighbors is guaranteed to be sorted.""" 649 if not (1 <= u <= self.lorder): 650 raise ValueError("Invalid choice of vertex") 651 return self.ladj.get(u, [])[:] 652 653 def left_neighbors(self, v): 654 """Outputs the neighbors of right vertex `u` 655 656 The sequence of neighbors is guaranteed to be sorted.""" 657 if not (1 <= v <= self.rorder): 658 raise ValueError("Invalid choice of vertex") 659 return self.radj.get(v, [])[:] 660 661 @classmethod 662 def from_networkx(cls, G): 663 """Convert a :py:class:`networkx.Graph` into a :py:class:`cnfgen.graphs.BipartiteGraph` 664 665 In order to convert a :py:class:`networkx.Graph` object `G`, 666 it is necessary that all nodes in `G` have the property 667 `bipartite` set to either `0` or `1`. 668 669 If this is not the case, or if there are edges between the two 670 parts, :py:class:`ValueError` is raised. 671 672 Example 673 ------- 674 >>> G = networkx.bipartite.complete_bipartite_graph(5,7) 675 >>> B = BipartiteGraph.from_networkx(G) 676 >>> print(B.order()) 677 12 678 >>> print(B.left_order()) 679 5 680 >>> print(B.has_edge(2,3)) 681 True 682 """ 683 if not isinstance(G, networkx.Graph): 684 raise ValueError('G is expected to be of type networkx.Graph') 685 side = [[], []] 686 index = [{}, {}] 687 for u in G.nodes(): 688 try: 689 color = G.nodes[u]['bipartite'] 690 assert color in ['0', 0, '1', 1] 691 except (KeyError, AssertionError): 692 raise ValueError( 693 "Node {} lacks the 'bipartite' property set to 0 or 1".format(u)) 694 side[int(color)].append(u) 695 696 B = cls(len(side[0]), len(side[1])) 697 index[0] = {u: i for (i, u) in enumerate(side[0], start=1)} 698 index[1] = {v: i for (i, v) in enumerate(side[1], start=1)} 699 for u, v in G.edges(): 700 ucolor = 0 if (u in index[0]) else 1 701 vcolor = 1 if (v in index[1]) else 0 702 703 if ucolor == vcolor: 704 raise ValueError( 705 "Edge ({},{}) across the bipartition".format(u, v)) 706 707 iu, iv = index[ucolor][u], index[vcolor][v] 708 if ucolor == 0: 709 B.add_edge(iu, iv) 710 else: 711 B.add_edge(iv, iu) 712 try: 713 B.name = G.name 714 except AttributeError: 715 B.name = '<unknown graph>' 716 return B 717 718 @classmethod 719 def graph_type_name(cls): 720 """Bipartite graphs are laleled as 'bipartite'""" 721 return 'bipartite' 722 723 @classmethod 724 def supported_file_formats(cls): 725 """File formats supported for bipartite graph I/O""" 726 if has_dot_library(): 727 return ['kthlist', 'gml', 'dot', 'matrix'] 728 else: 729 return ['kthlist', 'gml', 'matrix'] 730 731 @classmethod 732 def normalize(cls, G, varname='G'): 733 """Guarantees a cnfgen.graphs.BipartiteGraph object 734 735 If the given graph `G` is a networkx.Graph object with a bipartition, 736 this method produces a CNFgen bipartite graph object, relabeling 737 vertices so that vertices og each side are labeled as numbers from 1 738 to `n` and 1 to `m` respectively, where `n` and `m` are the numbers of 739 vertices in `G` on the left and right side, respectively. If the 740 vertices in the original graph have some kind of order, the order 741 is preserved. 742 743 If `G` is already a `cnfgen.graphs.BipartiteGraph` object, nothing is done. 744 745 """ 746 typemsg = "type of argument '{}' must be either networx.Graph or cnfgen.BipartiteGraph" 747 conversionmsg = "cannot convert '{}' to a bipartite graph: inconsistent 'bipartite' labeling" 748 if not isinstance(G, (BipartiteGraph, networkx.Graph)): 749 raise TypeError(typemsg.format(varname)) 750 if isinstance(G, BipartiteGraph): 751 return G 752 try: 753 G2 = cls.from_networkx(G) 754 return G2 755 except AttributeError: 756 raise ValueError(conversionmsg.format(varname)) 757 758 759 class CompleteBipartiteGraph(BipartiteGraph): 760 def __init__(self, L, R): 761 BipartiteGraph.__init__(self, L, R) 762 self.name = 'Complete bipartite graph with ({},{}) vertices'.format( 763 L, R) 764 765 def has_edge(self, u, v): 766 return (1 <= u <= self.lorder and 1 <= v <= self.rorder) 767 768 def add_edge(self, u, v): 769 pass 770 771 def number_of_edges(self): 772 return self.lorder * self.rorder 773 774 def right_neighbors(self, u): 775 return range(1, self.rorder + 1) 776 777 def left_neighbors(self, v): 778 return range(1, self.lorder + 1) 779 780 781 def has_dot_library(): 782 """Test the presence of pydot 783 """ 784 try: 785 # newer version of networkx 786 from networkx import nx_pydot 787 import pydot 788 del pydot 789 return True 790 except ImportError: 791 pass 792 793 return False 794 795 796 ################################################################# 797 # Graph reader/writer 798 ################################################################# 799 800 801 def guess_fileformat(fileorname, fileformat=None): 802 """Guess the file format for the file or filename """ 803 if fileformat is not None: 804 return fileformat 805 806 try: 807 if isinstance(fileorname, str): 808 name = fileorname 809 else: 810 name = fileorname.name 811 return os.path.splitext(name)[-1][1:] 812 except (AttributeError, ValueError, IndexError): 813 raise ValueError( 814 "Cannot guess a file format from arguments. Please specify the format manually.") 815 816 817 def _process_graph_io_arguments(iofile, graph_type, file_format, multi_edges): 818 """Test if the argument for the graph I/O functions make sense""" 819 820 # Check the file 821 if not isinstance(iofile, io.TextIOBase) and \ 822 not isinstance(iofile, io.IOBase) and \ 823 not isinstance(iofile, StringIO): 824 raise ValueError( 825 "The IO stream \"{}\" does not correspond to a file".format( 826 iofile)) 827 828 # Check the graph type specification 829 if graph_type not in ['dag', 'digraph', 'simple', 'bipartite']: 830 raise ValueError("The graph type must be one of " + 831 list(_graphformats.keys())) 832 833 if multi_edges: 834 raise NotImplementedError("Multi edges not supported yet") 835 836 elif graph_type in ["dag", "digraph"]: 837 grtype = DirectedGraph 838 elif graph_type == "simple": 839 grtype = Graph 840 elif graph_type == "bipartite": 841 grtype = BipartiteGraph 842 else: 843 raise RuntimeError( 844 "Unknown graph type argument: {}".format(graph_type)) 845 846 # Check/discover file format specification 847 if file_format == 'autodetect': 848 try: 849 extension = os.path.splitext(iofile.name)[-1][1:] 850 except AttributeError: 851 raise ValueError( 852 "Cannot guess a file format from an IO stream with no name. Please specify the format manually." 853 ) 854 if extension not in grtype.supported_file_formats(): 855 raise ValueError("Cannot guess a file format for {} graphs from the extension of \"{}\". Please specify the format manually.". 856 format(graph_type, iofile.name)) 857 else: 858 file_format = extension 859 860 elif file_format not in grtype.supported_file_formats(): 861 raise ValueError( 862 "For {} graphs we only support these formats: {}".format( 863 graph_type, grtype.supported_file_formats())) 864 865 return (grtype, file_format) 866 867 868 def normalize_networkx_labels(G): 869 """Relabel all vertices as integer starting from 1""" 870 # Normalize GML file. All nodes are integers starting from 1 871 try: 872 G = networkx.convert_node_labels_to_integers( 873 G, first_label=1, ordering='sorted') 874 except TypeError: 875 # Ids cannot be sorted natively 876 G = networkx.convert_node_labels_to_integers( 877 G, first_label=1, ordering='default') 878 return G 879 880 881 def readGraph(input_file, 882 graph_type, 883 file_format='autodetect', 884 multi_edges=False): 885 """Read a Graph from file 886 887 In the case of "bipartite" type, the graph obtained is of 888 :py:class:`cnfgen.graphs.BipartiteGraph`. 889 890 In the case of "simple" type, the graph is obtained of 891 :py:class:`cnfgen.graphs.Graph`. 892 893 In the case of "dag" or "directed" type, the graph obtained is of 894 :py:class:`cnfgen.graphs.DirectedGraph`. 895 896 The supported file formats are enumerated by the respective class method 897 ``supported_file_formats`` 898 899 In the case of "dag" type, the graph read in input must have 900 increasing edges, in the sense that all edges must be such that 901 the source has lower identifier than the sink. (I.e. the numeric 902 identifiers of the vertices are a topological order for the 903 graph) 904 905 Parameters 906 ----------- 907 input_file: str or file-like object 908 the input file from which the graph is read. If it is a string 909 then the graph is read from a file with that string as 910 filename. Otherwise if the input_file is a file object (or 911 a text stream), the graph is read from there. 912 913 Input files are assumed to be UTF-8 by default. 914 915 graph_type: string in {"simple","digraph","dag","bipartite"} 916 917 file_format: string, optional 918 The file format that the parser should expect to receive. 919 See also the method py:method::``supported_file_formats``. By default 920 it tries to autodetect it from the file name extension (when applicable). 921 922 multi_edges: bool,optional 923 are multiple edge allowed in the graph? By default this is not allowed. 924 925 Returns 926 ------- 927 a graph object 928 one type among Graph, DirectedGraph, BipartiteGraph 929 930 Raises 931 ------ 932 ValueError 933 raised when either ``input_file`` is neither a file object 934 nor a string, or when ``graph_type`` and ``file_format`` are 935 invalid choices. 936 937 IOError 938 it is impossible to read the ``input_file`` 939 940 See Also 941 -------- 942 writeGraph, is_dag, has_bipartition 943 944 """ 945 if multi_edges: 946 raise NotImplementedError("Multi edges not supported yet") 947 948 # file name instead of file object 949 if isinstance(input_file, str): 950 with open(input_file, 'r', encoding='utf-8') as file_handle: 951 return readGraph(file_handle, graph_type, file_format, multi_edges) 952 953 graph_class, file_format = _process_graph_io_arguments(input_file, 954 graph_type, 955 file_format, 956 multi_edges) 957 958 if file_format == 'dot': 959 960 # This is a workaround. In theory a broken dot file should 961 # cause a pyparsing.ParseError but the dot_reader used by 962 # networkx seems to mismanage that and to cause a TypeError 963 # 964 try: 965 G = networkx.nx_pydot.read_dot(input_file) 966 try: 967 # work around for a weird parse error in pydot, which 968 # adds an additiona vertex '\\n' in the graph. 969 G.remove_node('\\n') 970 except networkx.exception.NetworkXError: 971 pass 972 G = graph_class.normalize(G) 973 except TypeError: 974 raise ValueError('Parse Error in dot file') 975 976 elif file_format == 'gml': 977 978 # Networkx's GML reader expects to read from ascii encoded 979 # binary file. We could have sent the data to a temporary 980 # binary buffer but for some reasons networkx's GML reader 981 # function is poorly written and does not like such buffers. 982 # It turns out we can pass the data as a list of 983 # encoded ascii lines. 984 # 985 # The 'id' field in the vertices are supposed to be an integer 986 # and will be used as identifiers for the vertices in Graph 987 # object too. 988 # 989 try: 990 G = networkx.read_gml((line.encode('ascii') 991 for line in input_file), label='id') 992 G = graph_class.normalize(G) 993 except networkx.NetworkXError as errmsg: 994 raise ValueError("[Parse error in GML input] {} ".format(errmsg)) 995 except UnicodeEncodeError as errmsg: 996 raise ValueError( 997 "[Non-ascii chars in GML file] {} ".format(errmsg)) 998 999 elif file_format == 'kthlist' and graph_type == 'bipartite': 1000 1001 G = _read_bipartite_kthlist(input_file) 1002 1003 elif file_format == 'kthlist' and graph_type != 'bipartite': 1004 1005 G = _read_nonbipartite_kthlist(input_file, graph_class) 1006 1007 elif file_format == 'dimacs': 1008 1009 G = _read_graph_dimacs_format(input_file, graph_class) 1010 1011 elif file_format == 'matrix': 1012 1013 G = _read_graph_matrix_format(input_file) 1014 1015 else: 1016 raise RuntimeError( 1017 "[Internal error] Format {} not implemented".format(file_format)) 1018 1019 if graph_type == "dag" and not G.is_dag(): 1020 raise ValueError( 1021 "[Input error] Graph must be explicitly acyclic (src->dest edges where src<dest)") 1022 1023 return G 1024 1025 1026 def writeGraph(G, output_file, graph_type, file_format='autodetect'): 1027 """Write a graph to a file 1028 1029 Parameters 1030 ----------- 1031 G : BaseGraph 1032 1033 output_file: file object 1034 the output file to which the graph is written. If it is a string 1035 then the graph is written to a file with that string as 1036 filename. Otherwise if ``output_file`` is a file object (or 1037 a text stream), the graph is written there. 1038 1039 The file is written in UTF-8 by default. 1040 1041 graph_type: string in {"simple","digraph","dag","bipartite"} 1042 see also :py:func:`cnfgen.graph.supported_formats` 1043 1044 file_format: string, optional 1045 The file format that the parser should expect to receive. 1046 See also :py:func:`cnfgen.graph.supported_formats`. By default 1047 it tries to autodetect it from the file name extension (when applicable). 1048 1049 Returns 1050 ------- 1051 None 1052 1053 Raises 1054 ------ 1055 ValueError 1056 raised when either ``output_file`` is neither a file object 1057 nor a string, or when ``graph_type`` and ``file_format`` are 1058 invalid choices. 1059 1060 IOError 1061 it is impossible to write on the ``output_file`` 1062 1063 See Also 1064 -------- 1065 readGraph 1066 1067 """ 1068 if not isinstance(G, BaseGraph): 1069 raise TypeError("G must be a cnfgen.graphs.BaseGraph") 1070 1071 # file name instead of file object 1072 if isinstance(output_file, str): 1073 with open(output_file, 'w', encoding='utf-8') as file_handle: 1074 return writeGraph(G, file_handle, graph_type, file_format) 1075 1076 _, file_format = _process_graph_io_arguments(output_file, graph_type, 1077 file_format, False) 1078 1079 if file_format == 'dot': 1080 1081 G = G.to_networkx() 1082 networkx.nx_pydot.write_dot(G, output_file) 1083 1084 elif file_format == 'gml': 1085 1086 # Networkx's GML writer expects to write to an ascii encoded 1087 # binary file. Thus we need to let Networkx write to 1088 # a temporary binary ascii encoded buffer and then convert the 1089 # content before sending it to the output file. 1090 tempbuffer = io.BytesIO() 1091 G = G.to_networkx() 1092 networkx.write_gml(G, tempbuffer) 1093 print(tempbuffer.getvalue().decode('ascii'), file=output_file) 1094 1095 elif file_format == 'kthlist' and graph_type != 'bipartite': 1096 1097 _write_graph_kthlist_nonbipartite(G, output_file) 1098 1099 elif file_format == 'kthlist' and graph_type == 'bipartite': 1100 1101 _write_graph_kthlist_bipartite(G, output_file) 1102 1103 elif file_format == 'dimacs': 1104 1105 _write_graph_dimacs_format(G, output_file) 1106 1107 elif file_format == 'matrix': 1108 1109 _write_graph_matrix_format(G, output_file) 1110 1111 else: 1112 raise RuntimeError( 1113 "[Internal error] Format {} not implemented".format(file_format)) 1114 1115 1116 # 1117 # In-house parsers 1118 # 1119 def _kthlist_parse(inputfile): 1120 """Read a graph from file, and produce the datas. 1121 1122 First yeild (#vertex,first comment line) 1123 Then generates a sequence of (s,target,lineno) 1124 1125 Raises: 1126 ValueError is parsing fails for some reason 1127 """ 1128 # vertex number 1129 size = -1 1130 name = "" 1131 1132 for i, l in enumerate(inputfile.readlines()): 1133 1134 # first non empty comment line is the graph name 1135 # must be before the graph size 1136 if l[0] == 'c': 1137 if size < 0 and len(name) == 0 and len(l[2:].strip()) != 0: 1138 name += l[2:] 1139 continue 1140 1141 # empty line 1142 if len(l.strip()) == 0: 1143 continue 1144 1145 if ':' not in l: 1146 # vertex number spec 1147 if size >= 0: 1148 raise ValueError( 1149 "Line {} contains a second spec directive.".format(i)) 1150 try: 1151 size = int(l.strip()) 1152 if size < 0: 1153 raise ValueError 1154 except ValueError: 1155 raise ValueError( 1156 "Non negative number expected at line {}.".format(i)) 1157 yield (size, name) 1158 continue 1159 1160 # Load edges from this line 1161 left, right = l.split(':') 1162 try: 1163 left = int(left.strip()) 1164 right = [int(s) for s in right.split()] 1165 except ValueError: 1166 raise ValueError("Non integer vertex ID at line {}.".format(i)) 1167 if len(right) < 1 or right[-1] != 0: 1168 raise ValueError("Line {} must end with 0.".format(i)) 1169 1170 if left < 1 or left > size: 1171 raise ValueError( 1172 "Vertex ID out of range [1,{}] at line {}.".format(size, i)) 1173 1174 right.pop() 1175 if len([x for x in right if x < 1 or x > size]) > 0: 1176 raise ValueError( 1177 "Vertex ID out of range [1,{}] at line {}.".format(size, i)) 1178 yield left, right, i 1179 1180 1181 def _read_bipartite_kthlist(inputfile): 1182 """Read a bipartite graph from file, in the KTH reverse adjacency lists format. 1183 1184 Assumes the adjacecy list is given in order. 1185 - vertices are listed in increasing order 1186 - if bipartite, only the adjiacency list of the left side must be 1187 given, no list for a vertex of the right side is allowed. 1188 1189 Parameters 1190 ---------- 1191 inputfile : file object 1192 file handle of the input 1193 1194 Raises 1195 ------ 1196 ValueError 1197 Error parsing the file 1198 1199 """ 1200 # vertex number 1201 parser = _kthlist_parse(inputfile) 1202 size, name = next(parser) 1203 bipartition_ambiguous = [1, size] 1204 edges = {} 1205 1206 previous = 0 1207 for left, right, lineno in parser: 1208 1209 if left <= previous: 1210 raise ValueError( 1211 "Vertex at line {} is smaller than the previous one.".format(lineno)) 1212 1213 # Check the bi-coloring on both side 1214 if left > bipartition_ambiguous[1]: 1215 raise ValueError( 1216 "Bipartition violation al line {}. Vertex {} cannot be on the left side." 1217 .format(lineno, left)) 1218 bipartition_ambiguous[0] = max(bipartition_ambiguous[0], left + 1) 1219 for v in right: 1220 if v < bipartition_ambiguous[0]: 1221 raise ValueError( 1222 "Bipartition violation. Invalid edge ({},{}) at line {}." 1223 .format(left, v, lineno)) 1224 bipartition_ambiguous[1] = min(bipartition_ambiguous[1], v - 1) 1225 1226 # after vertices, add the edges 1227 edges[left] = right 1228 1229 # fix the bipartition 1230 # unsassigned vertices go to the right size 1231 L = bipartition_ambiguous[0]-1 1232 R = size - bipartition_ambiguous[0]+1 1233 G = BipartiteGraph(L, R, name) 1234 1235 for u in edges: 1236 for v in edges[u]: 1237 G.add_edge(u, v - L) 1238 1239 if size != G.number_of_vertices(): 1240 raise ValueError("{} vertices expected. Got {} instead.".format( 1241 size, G.number_of_vertices())) 1242 return G 1243 1244 1245 def _read_nonbipartite_kthlist(inputfile, graph_class): 1246 """Read a graph from file, in the KTH reverse adjacency lists format. 1247 1248 Only for simple and directed graph 1249 1250 Assumes the adjacecy list is given in order. 1251 - vertices are listed in increasing order 1252 - if directed graph the adjacency list specifies incoming neighbous 1253 - if DAG, the graph must be given in topological order source->sink 1254 1255 Parameters 1256 ---------- 1257 inputfile : file object 1258 file handle of the input 1259 1260 graph_class: class 1261 either Graph or DirectedGraph 1262 1263 Raises 1264 ------ 1265 ValueError 1266 Error parsing the file 1267 1268 """ 1269 assert graph_class in [Graph, DirectedGraph] 1270 1271 # vertex number 1272 parser = _kthlist_parse(inputfile) 1273 size, name = next(parser) 1274 G = graph_class(size, name) 1275 1276 previous = 0 1277 for succ, predecessors, lineno in parser: 1278 1279 if succ <= previous: 1280 raise ValueError( 1281 "Vertex at line {} is smaller than the previous one.".format(lineno)) 1282 1283 # after vertices, add the edges 1284 for v in predecessors: 1285 G.add_edge(v, succ) 1286 1287 previous = succ 1288 1289 if size != G.order(): 1290 raise ValueError("{} vertices expected. Got {} instead.".format( 1291 size, G.order())) 1292 1293 return G 1294 1295 1296 def _read_graph_dimacs_format(inputfile, graph_class): 1297 """Read a graph simple from file, in the DIMACS edge format. 1298 1299 Parameters 1300 ---------- 1301 inputfile : file object 1302 file handle of the input 1303 1304 graph_class: class object 1305 either Graph or DirectedGraph 1306 """ 1307 assert graph_class in [Graph, DirectedGraph] 1308 1309 G = None 1310 name = '' 1311 n = -1 1312 m = -1 1313 m_cnt = 0 1314 1315 # is the input topologically sorted? 1316 for i, l in enumerate(inputfile.readlines()): 1317 1318 l = l.strip() 1319 1320 # add the comment to the header 1321 if l[0] == 'c': 1322 name += l[2:] 1323 continue 1324 1325 # parse spec line 1326 if l[0] == 'p': 1327 if G is not None: 1328 raise ValueError( 1329 "[Syntax error] " + 1330 "Line {} contains a second spec line.".format(i+1)) 1331 _, fmt, nstr, mstr = l.split() 1332 if fmt != 'edge': 1333 raise ValueError("[Input error] " + 1334 "Dimacs \'edge\' format expected at line {}.".format(i+1)) 1335 n = int(nstr) 1336 m = int(mstr) 1337 G = graph_class(n, name) 1338 continue 1339 1340 # parse spec line 1341 if l[0] == 'e': 1342 if G is None: 1343 raise ValueError("[Input error] " + 1344 "Edge before preamble at line".format(i)) 1345 m_cnt += 1 1346 _, v, w = l.split() 1347 try: 1348 G.add_edge(int(v), int(w)) 1349 except ValueError: 1350 raise ValueError("[Syntax error] " + 1351 "Line {} syntax error: edge must be 'e u v' where u, v are vertices".format(i)) 1352 1353 if m != m_cnt: 1354 raise ValueError("[Syntax error] " + 1355 "{} edges were expected.".format(m)) 1356 1357 return G 1358 1359 1360 def _read_graph_matrix_format(inputfile): 1361 """Read a bipartite graph from file, in the adjiacency matrix format. 1362 1363 This is an example of an adjacency matrix for a bipartite graph 1364 with 9 vertices on one side and 15 on the another side. 1365 1366 .. 9 15 1367 1 1 0 1 0 0 0 1 0 0 0 0 0 0 0 1368 0 1 1 0 1 0 0 0 1 0 0 0 0 0 0 1369 0 0 1 1 0 1 0 0 0 1 0 0 0 0 0 1370 0 0 0 1 1 0 1 0 0 0 1 0 0 0 0 1371 0 0 0 0 1 1 0 1 0 0 0 1 0 0 0 1372 0 0 0 0 0 1 1 0 1 0 0 0 1 0 0 1373 0 0 0 0 0 0 1 1 0 1 0 0 0 1 0 1374 0 0 0 0 0 0 0 1 1 0 1 0 0 0 1 1375 1 0 0 0 0 0 0 0 1 1 0 1 0 0 0 1376 1377 Parameters 1378 ---------- 1379 inputfile: file object 1380 the file containing the graph specification 1381 1382 Returns 1383 ------- 1384 G : BipartiteGraph 1385 1386 """ 1387 def scan_integer(inputfile): 1388 1389 num_buffer = [] 1390 line_cnt = 0 1391 1392 while True: 1393 if len(num_buffer) == 0: 1394 1395 line = inputfile.readline() 1396 1397 if len(line) == 0: 1398 return 1399 1400 line_cnt += 1 1401 tokens = line.split() 1402 1403 if len(tokens) == 0 or tokens[0][0] == '#': 1404 continue # comment line 1405 1406 try: 1407 num_buffer.extend((int(lit), line_cnt) for lit in tokens) 1408 except ValueError: 1409 raise ValueError("[Syntax error] " + 1410 "Line {} contains a non numeric entry.". 1411 format(line_cnt)) 1412 1413 yield num_buffer.pop(0) 1414 1415 scanner = scan_integer(inputfile) 1416 1417 try: 1418 n = next(scanner)[0] 1419 m = next(scanner)[0] 1420 1421 G = BipartiteGraph(n, m) 1422 G.name = '' 1423 1424 # read edges 1425 for i in range(1, n + 1): 1426 for j in range(1, m + 1): 1427 1428 (b, l) = next(scanner) 1429 if b == 1: 1430 G.add_edge(i, j) 1431 elif b == 0: 1432 pass 1433 else: 1434 raise ValueError( 1435 "[Input error at line {}] Only 0 or 1 are allowed". 1436 format(l)) 1437 except StopIteration: 1438 raise ValueError("[Input error] Unexpected end of the matrix") 1439 1440 # check that there are is no more data 1441 try: 1442 (b, l) = next(scanner) 1443 raise ValueError( 1444 "[Input error at line {}] There are more than {}x{} entries". 1445 format(l, n, m)) 1446 except StopIteration: 1447 pass 1448 1449 return G 1450 1451 1452 # 1453 # In-house graph writers 1454 # 1455 def _write_graph_kthlist_nonbipartite(G, output_file): 1456 """Wrire a graph to a file, in the KTH reverse adjacency lists format. 1457 1458 Parameters 1459 ---------- 1460 G : Graph or DirectGraph 1461 the graph to write on file 1462 1463 output_file : file object 1464 file handle of the output 1465 """ 1466 assert isinstance(G, (Graph, DirectedGraph)) 1467 1468 print("c {}".format(G.name), file=output_file) 1469 print("{}".format(G.order()), file=output_file) 1470 1471 from io import StringIO 1472 output = StringIO() 1473 1474 for v in G.vertices(): 1475 1476 if G.is_directed(): 1477 nbors = G.predecessors(v) 1478 else: 1479 nbors = G.neighbors(v) 1480 1481 output.write(str(v) + " :") 1482 output.write("".join([' '+str(i) for i in nbors])) 1483 output.write(" 0\n") 1484 1485 print(output.getvalue(), file=output_file) 1486 1487 1488 def _write_graph_kthlist_bipartite(G, output_file): 1489 """Wrire a bipartite graph to a file, 1490 in the KTH reverse adjacency lists format. 1491 1492 Parameters 1493 ---------- 1494 G : BipartiteGraph 1495 the graph to write on file 1496 1497 output_file : file object 1498 file handle of the output 1499 """ 1500 assert isinstance(G, BipartiteGraph) 1501 print("c {}".format(G.name), file=output_file) 1502 print("{}".format(G.order()), file=output_file) 1503 1504 from io import StringIO 1505 output = StringIO() 1506 1507 U, _ = G.parts() 1508 offset = len(U) 1509 1510 for u in U: 1511 output.write(str(u) + " :") 1512 output.write("".join([' '+str(v + offset) 1513 for v in G.right_neighbors(u)])) 1514 output.write(" 0\n") 1515 1516 print(output.getvalue(), file=output_file) 1517 1518 1519 def _write_graph_dimacs_format(G, output_file): 1520 """Wrire a graph to a file, in DIMACS format. 1521 1522 Parameters 1523 ---------- 1524 G : Graph or DirectGraph 1525 the graph to write on file 1526 1527 output_file : file object 1528 file handle of the output 1529 """ 1530 assert isinstance(G, (Graph, DirectedGraph)) 1531 print("c {}".format(G.name).strip(), file=output_file) 1532 n = G.number_of_vertices() 1533 m = G.number_of_edges() 1534 print("p edge {} {}".format(n, m), file=output_file) 1535 1536 for v, w in G.edges(): 1537 print("e {} {}".format(v, w), file=output_file) 1538 1539 1540 def _write_graph_matrix_format(G, output_file): 1541 """Wrire a graph to a file, in \"matrix\" format. 1542 1543 Parameters 1544 ---------- 1545 G : BipartiteGraph 1546 the graph to write in output 1547 1548 output_file : file object 1549 file handle of the output 1550 """ 1551 assert isinstance(G, BipartiteGraph) 1552 print("{} {}".format(G.left_order(), G.right_order()), 1553 file=output_file) 1554 L, R = G.parts() 1555 for u in L: 1556 1557 adj_row = [] 1558 1559 for v in R: 1560 if G.has_edge(u, v): 1561 adj_row.append("1") 1562 else: 1563 adj_row.append("0") 1564 1565 print(" ".join(adj_row), file=output_file) 1566 1567 1568 # 1569 # Bipartite graph generator 1570 # (we do not want to use networkx) 1571 # 1572 def bipartite_random_left_regular(l, r, d, seed=None): 1573 """Returns a random bipartite graph with constant left degree. 1574 1575 Each vertex on the left side has `d` neighbors on the right side, 1576 picked uniformly at random without repetition. 1577 1578 Each vertex in the graph has an attribute `bipartite` which is 0 1579 for the vertices on the left side and 1 for the vertices on the 1580 right side. 1581 1582 Parameters 1583 ---------- 1584 l : int 1585 vertices on the left side 1586 r : int 1587 vertices on the right side 1588 d : int 1589 degree on the left side. 1590 seed : hashable object 1591 seed the random generator 1592 1593 Returns 1594 ------- 1595 BipartiteGraph 1596 1597 Raises 1598 ------ 1599 ValueError 1600 unless ``l``, ``r`` and ``d`` are non negative. 1601 1602 """ 1603 import random 1604 if seed is not None: 1605 random.seed(seed) 1606 1607 if l < 0 or r < 0 or d < 0: 1608 raise ValueError( 1609 "bipartite_random_left_regular(l,r,d) needs l,r,d >=0.") 1610 1611 G = BipartiteGraph(l, r) 1612 G.name = "bipartite_random_left_regular({},{},{})".format(l, r, d) 1613 d = min(r, d) 1614 1615 L, R = G.parts() 1616 for u in L: 1617 for v in sorted(random.sample(R, d)): 1618 G.add_edge(u, v) 1619 1620 return G 1621 1622 1623 def bipartite_random_m_edges(L, R, m, seed=None): 1624 """Returns a random bipartite graph with M edges 1625 1626 Build a random bipartite graph with :math:`L` left vertices, 1627 :math:`R` right vertices and :math:`m` edges sampled at random 1628 without repetition. 1629 1630 Parameters 1631 ---------- 1632 L : int 1633 vertices on the left side 1634 R : int 1635 vertices on the right side 1636 m : int 1637 number of edges. 1638 seed : hashable object 1639 seed the random generator 1640 1641 Returns 1642 ------- 1643 BipartiteGraph 1644 1645 Raises 1646 ------ 1647 ValueError 1648 unless ``L``, ``R`` and ``m`` are non negative. 1649 1650 """ 1651 import random 1652 if seed is not None: 1653 random.seed(seed) 1654 1655 if L < 1 or R < 1 or m < 0 or m > L * R: 1656 raise ValueError( 1657 "bipartite_random_m_edges(L,R,m) needs L, R >= 1, 0<=m<=L*R") 1658 G = BipartiteGraph(L, R) 1659 G.name = "bipartite_random_m_edges({},{},{})".format(L, R, m) 1660 1661 U, V = G.parts() 1662 1663 if m > L * R // 3: 1664 # Sampling strategy (dense) 1665 E = ((u, v) for u in U for v in V) 1666 for u, v in random.sample(E, m): 1667 G.add_edge(u, v) 1668 else: 1669 # Sampling strategy (sparse) 1670 count = 0 1671 while count < m: 1672 u = random.randint(1, L) 1673 v = random.randint(1, R) 1674 if not G.has_edge(u, v): 1675 G.add_edge(u, v) 1676 count += 1 1677 assert G.number_of_edges() == m 1678 return G 1679 1680 1681 def bipartite_random(L, R, p, seed=None): 1682 """Returns a random bipartite graph with independent edges 1683 1684 Build a random bipartite graph with :math:`L` left vertices, 1685 :math:`R` right vertices, where each edge is sampled independently 1686 with probability :math:`p`. 1687 1688 Parameters 1689 ---------- 1690 L : int 1691 vertices on the left side 1692 R : int 1693 vertices on the right side 1694 p : float 1695 probability to pick an edge 1696 seed : hashable object 1697 seed the random generator 1698 1699 Returns 1700 ------- 1701 BipartiteGraph 1702 1703 Raises 1704 ------ 1705 ValueError 1706 unless ``L``, ``R`` are non negative and 0<=``p``<=1. 1707 """ 1708 import random 1709 if seed is not None: 1710 random.seed(seed) 1711 1712 if L < 1 or R < 1 or p < 0 or p > 1: 1713 raise ValueError( 1714 "bipartite_random_graph(L,R,p) needs L, R >= 1, p in [0,1]") 1715 G = BipartiteGraph(L, R) 1716 G.name = "bipartite_random_graph({},{},{})".format(L, R, p) 1717 1718 U, V = G.parts() 1719 1720 for u in U: 1721 for v in V: 1722 if random.random() <= p: 1723 G.add_edge(u, v) 1724 return G 1725 1726 1727 def bipartite_shift(N, M, pattern=[]): 1728 """Returns a bipartite graph where edges are a fixed shifted sequence. 1729 1730 The graph has :math:`N` vertices on the left (numbered from 1731 :math:`1` to :math:`N`), and :math:`M` vertices on the right 1732 (numbered from :math:`1` to :math:`M`), 1733 1734 Each vertex :math:`v` on the left side has edges to vertices 1735 :math:`v+d_1`, :math:`v+d_2`, :math:`v+d_3`,... with vertex 1736 indices on the right wrap around :wrap around over 1737 :math:`[1..M]`). 1738 1739 Notice that this construction does not produces multiedges even if 1740 two offsets end up on the same right vertex. 1741 1742 Parameters 1743 ---------- 1744 N : int 1745 vertices on the left side 1746 M : int 1747 vertices on the right side 1748 pattern : list(int) 1749 pattern of neighbors 1750 1751 Returns 1752 ------- 1753 BipartiteGraph 1754 1755 Raises 1756 ------ 1757 ValueError 1758 unless ``N``, ``M`` are non negative and ``pattern`` has vertices outside the range. 1759 1760 """ 1761 if N < 1 or M < 1: 1762 raise ValueError("bipartite_shift(N,M,pattern) needs N,M >= 0.") 1763 1764 G = BipartiteGraph(N, M) 1765 G.name = "bipartite_shift_regular({},{},{})".format(N, M, pattern) 1766 1767 L, R = G.parts() 1768 pattern.sort() 1769 for u in L: 1770 for offset in pattern: 1771 G.add_edge(u, 1 + (u - 1 + offset) % M) 1772 1773 return G 1774 1775 1776 def bipartite_random_regular(l, r, d, seed=None): 1777 """Returns a random bipartite graph with constant degree on both sides. 1778 1779 The graph is d-regular on the left side and regular on the right 1780 size, so it must be that d*l / r is an integer number. 1781 1782 Parameters 1783 ---------- 1784 l : int 1785 vertices on the left side 1786 r : int 1787 vertices on the right side 1788 d : int 1789 degree of vertices at the left side 1790 seed : hashable object 1791 seed of random generator 1792 1793 Returns 1794 ------- 1795 BipartiteGraph 1796 1797 Raises 1798 ------ 1799 ValueError 1800 if one among ``l``, ``r`` and ``d`` is negative or 1801 if ``r`` does not divides `l*d` 1802 1803 References 1804 ---------- 1805 [1] http://... 1806 1807 """ 1808 1809 import random 1810 if seed is not None: 1811 random.seed(seed) 1812 1813 if l < 0 or r < 0 or d < 0: 1814 raise ValueError("bipartite_random_regular(l,r,d) needs l,r,d >=0.") 1815 1816 if (l * d) % r != 0: 1817 raise ValueError( 1818 "bipartite_random_regular(l,r,d) needs r to divid l*d.") 1819 1820 G = BipartiteGraph(l, r) 1821 G.name = "bipartite_random_regular({},{},{})".format(l, r, d) 1822 1823 L, R = G.parts() 1824 A = list(L) * d 1825 B = list(R) * (l * d // r) 1826 assert len(B) == l * d 1827 1828 for i in range(l * d): 1829 # Sample an edge, do not add it if it existed 1830 # We expect to sample at most d^2 edges 1831 for retries in range(3 * d * d): 1832 ea = random.randint(i, l * d - 1) 1833 eb = random.randint(i, l * d - 1) 1834 if not G.has_edge(A[ea], B[eb]): 1835 G.add_edge(A[ea], B[eb]) 1836 A[i], A[ea] = A[ea], A[i] 1837 B[i], B[eb] = B[eb], B[i] 1838 break 1839 else: 1840 # Sampling takes too long, maybe no good edge exists 1841 failure = True 1842 for ea in range(i, l * d): 1843 for eb in range(i, l * d): 1844 if not G.has_edge(A[ea], B[eb]): 1845 failure = False 1846 break 1847 if not failure: 1848 break 1849 if failure: 1850 return bipartite_random_regular(l, r, d) 1851 1852 return G 1853 1854 1855 def dag_pyramid(height): 1856 """Generates the pyramid DAG 1857 1858 Vertices are indexed from the bottom layer, starting from index 1 1859 1860 Parameters 1861 ---------- 1862 height : int 1863 the height of the pyramid graph (>=0) 1864 1865 Returns 1866 ------- 1867 cnfgen.graphs.DirectedGraph 1868 1869 Raises 1870 ------ 1871 ValueError 1872 """ 1873 if height < 0: 1874 raise ValueError("The height of the tree must be >= 0") 1875 1876 n = (height+1)*(height+2) // 2 # number of vertices 1877 D = DirectedGraph(n, 'Pyramid of height {}'.format(height)) 1878 1879 # edges 1880 leftsrc = 1 1881 dest = height+2 1882 for layer in range(1, height+1): 1883 for i in range(1, height-layer+2): 1884 D.add_edge(leftsrc, dest) 1885 D.add_edge(leftsrc+1, dest) 1886 leftsrc += 1 1887 dest += 1 1888 leftsrc += 1 1889 1890 return D 1891 1892 1893 def dag_complete_binary_tree(height): 1894 """Generates the complete binary tree DAG 1895 1896 Vertices are indexed from the bottom layer, starting from index 1 1897 1898 Parameters 1899 ---------- 1900 height : int 1901 the height of the tree 1902 1903 Returns 1904 ------- 1905 cnfgen.graphs.DirectedGraph 1906 1907 Raises 1908 ------ 1909 ValueError 1910 1911 """ 1912 if height < 0: 1913 raise ValueError("The height of the tree must be >= 0") 1914 1915 # vertices plus 1 1916 N = 2 * (2**height) 1917 name = 'Complete binary tree of height {}'.format(height) 1918 D = DirectedGraph(N-1, name) 1919 1920 # edges 1921 leftsrc = 1 1922 for dest in range(N // 2 + 1, N): 1923 D.add_edge(leftsrc, dest) 1924 D.add_edge(leftsrc+1, dest) 1925 leftsrc += 2 1926 1927 return D 1928 1929 1930 def dag_path(length): 1931 """Generates a directed path DAG 1932 1933 Vertices are indexed from 1..length+1 1934 1935 Parameters 1936 ---------- 1937 length : int 1938 the length of the path 1939 1940 Returns 1941 ------- 1942 cnfgen.graphs.DirectedGraph 1943 1944 Raises 1945 ------ 1946 ValueError 1947 """ 1948 if length < 0: 1949 raise ValueError("The lenght of the path must be >= 0") 1950 1951 name = 'Directed path of length {}'.format(length) 1952 D = DirectedGraph(length+1, name) 1953 # edges 1954 for i in range(1, length+1): 1955 D.add_edge(i, i + 1) 1956 1957 return D 1958 1959 1960 def split_random_edges(G,k, seed=None): 1961 """Split m random missing edges to G 1962 1963 If :math:`G` is a simple graph, it picks k random edges (and fails 1964 if there are not enough of them), and splits the edges in 2 adding 1965 a new vertex for each of them. 1966 1967 Parameters 1968 ---------- 1969 G : Graph 1970 a graph with at least :math:`m` missing edges 1971 k : int 1972 the number of edges to sample 1973 seed : hashable object 1974 seed of random generator 1975 1976 Example 1977 ------- 1978 >>> G = Graph(5) 1979 >>> G.add_edges_from([(1,4),(4,5),(2,4),(2,3)]) 1980 >>> G.number_of_edges() 1981 4 1982 >>> split_random_edges(G,2) 1983 >>> G.number_of_edges() 1984 6 1985 >>> G.number_of_vertices() 1986 7 1987 """ 1988 if seed is not None: 1989 random.seed(seed) 1990 1991 if not isinstance(G,Graph): 1992 raise TypeError("Edge splitting is only implemented for simple graphs") 1993 1994 non_negative_int(k,'k') 1995 if k > G.number_of_edges(): 1996 raise ValueError("The graph does not have {} edges.".format(k)) 1997 1998 tosplit = random.sample(list(G.edges()),k) 1999 nv = G.number_of_vertices() 2000 G.update_vertex_number(nv+k) 2001 x = nv + 1 2002 for u,v in tosplit: 2003 G.remove_edge(u,v) 2004 G.add_edge(u,x) 2005 G.add_edge(x,v) 2006 x += 1 2007 2008 2009 def add_random_missing_edges(G, m, seed=None): 2010 """Add m random missing edges to G 2011 2012 If :math:`G` is not complete and has at least :math:`m` missing 2013 edges, :math:`m` of them are sampled and added to the graph. 2014 2015 Parameters 2016 ---------- 2017 G : Graph 2018 a graph with at least :math:`m` missing edges 2019 m : int 2020 the number of missing edges to sample 2021 seed : hashable object 2022 seed of random generator 2023 2024 Raises 2025 ------ 2026 ValueError 2027 if :math:`G` doesn't have :math:`m` missing edges 2028 RuntimeError 2029 Sampling failure in the sparse case 2030 2031 """ 2032 if seed is not None: 2033 random.seed(seed) 2034 2035 if m < 0: 2036 raise ValueError("You can only sample a non negative number of edges.") 2037 2038 total_number_of_edges = None 2039 2040 if G.is_bipartite(): 2041 2042 Left, Right = G.parts() 2043 total_number_of_edges = len(Left) * len(Right) 2044 2045 def edge_sampler(): 2046 u = random.sample(Left, 1)[0] 2047 v = random.sample(Right, 1)[0] 2048 return (u, v) 2049 2050 def available_edges(): 2051 return [(u, v) for u in Left for v in Right if not G.has_edge(u, v)] 2052 2053 else: 2054 2055 V = G.number_of_vertices() 2056 total_number_of_edges = V * (V - 1) / 2 2057 2058 def edge_sampler(): 2059 return random.sample(range(1, V+1), 2) 2060 2061 def available_edges(): 2062 result = [] 2063 for u in range(1, V): 2064 for v in range(u+1, V+1): 2065 if not G.has_edge(u, v): 2066 result.append((u, v)) 2067 return result 2068 2069 # How many edges we want in the end? 2070 goal = G.number_of_edges() + m 2071 2072 if goal > total_number_of_edges: 2073 raise ValueError( 2074 "The graph does not have {} missing edges to sample.".format(m)) 2075 2076 # Sparse case: sample and retry 2077 for _ in range(10 * m): 2078 2079 if G.number_of_edges() >= goal: 2080 break 2081 2082 u, v = edge_sampler() 2083 if not G.has_edge(u, v): 2084 G.add_edge(u, v) 2085 2086 if G.number_of_edges() < goal: 2087 # Very unlikely case: sampling process failed and the solution 2088 # is to use the sampling process tailored for denser graph, so 2089 # that a correct result is guaranteed. This requires 2090 # generating all available edges 2091 for u, v in random.sample(available_edges(), 2092 goal - G.number_of_edges()): 2093 G.add_edge(u, v) 2094 2095 2096 def supported_graph_formats(): 2097 """File formats supported for graph I/O 2098 2099 Given as a dictionary that maps graph types to the respective 2100 supported formats. 2101 2102 E.g. 'dag' -> ['dimacs', 'kthlist'] 2103 """ 2104 return {'simple': Graph.supported_file_formats(), 2105 'digraph': DirectedGraph.supported_file_formats(), 2106 'dag': DirectedGraph.supported_file_formats(), 2107 'bipartite': BipartiteGraph.supported_file_formats()}
localtypes.py 1 #!/usr/bin/env python 2 # -*- coding:utf-8 -*- 3 """Functions to check the arguments types 4 """ 5 6 import numbers 7 8 9 def positive_int(value, name): 10 """Check that `value` is a positive integer""" 11 msg = "argument '{}' must be a positive integer".format(name) 12 if not isinstance(value, numbers.Integral): 13 raise TypeError(msg) 14 if value < 1: 15 raise ValueError(msg) 16 17 def positive_int_seq(value, name): 18 """Check that `value` is a positive integer""" 19 msg = "argument '{}' must be a sequence of positive integers".format(name) 20 try: 21 for v in value: 22 if not isinstance(v, numbers.Integral): 23 raise TypeError('non numeric element in the sequence') 24 except TypeError as te: 25 raise TypeError(msg) from te 26 27 for v in value: 28 if v < 1: 29 raise ValueError(msg) 30 31 def non_negative_int_seq(value, name): 32 """Check that `value` is a positive integer""" 33 msg = "argument '{}' must be a sequence of non negative integers".format(name) 34 try: 35 for v in value: 36 if not isinstance(v, numbers.Integral): 37 raise TypeError('non numeric element in the sequence') 38 except TypeError as te: 39 raise TypeError(msg) from te 40 41 for v in value: 42 if v < 0: 43 raise ValueError(msg) 44 45 def one_of_values(value, name, choices): 46 '''Check if the value is in a specific set''' 47 msg = "argument '{}' must be one of [{}]".format(name, 48 choices) 49 if value not in choices: 50 raise ValueError(msg) 51 52 def any_int(value, name): 53 """Check that `value` is an integer""" 54 msg = "argument '{}' must be have integer value".format(name) 55 if not isinstance(value, numbers.Integral): 56 raise TypeError(msg) 57 58 def non_negative_int(value, name): 59 """Check that the `value` is a non negative""" 60 msg = "argument '{}' must be a non negative integer".format(name) 61 if not isinstance(value, numbers.Integral): 62 raise TypeError(msg) 63 if value < 0: 64 raise ValueError(msg) 65 66 67 def probability_value(value, name): 68 """Check that the `value` is a real between 0 and 1""" 69 msg = "argument '{}' must be a real between 0 and 1".format(name) 70 if not isinstance(value, numbers.Real): 71 raise TypeError(msg) 72 if value < 0 or value > 1: 73 raise ValueError(msg)
2.1 Generating hard instances Considering the outline of methods used in current DPLL-based SAT-solvers we can now make an observation about graphs which should lead to hard instances EC(G). First let us restrict our attention to graphs G which are obtained by picking an edge in a 4-regular graph on n vertices and subdividing it, i.e. introducing a new vertex as midpoint of the old edge. For such a graph EC(G) will be unsatisfiable, have 2n + 1 variables and 10n + 2 clauses. This gives us unsatisfiable formulae with a density of close to 5 clauses per variable. For formulae with clauses of length 4 this is a density for which a formula from the random 4-SAT ensemble is expected to be satisfiable, see e.g. [1]. In this random model a formula is constructed by for each possible clause of length 4, for a set of n variables, letting it be part of the formula with probability p. Random 4-SAT formulae with expected density 5 are also expected to be easy to solve. 考虑到目前基于dpll的sat求解器所使用的方法的大纲,我们现在可以对图进行观察,这将导致硬实例EC(G)。 首先让我们将注意力限制在图G上,图G是通过在n个顶点上的4正则图中选取一条边并将其细分而得到的,即引入一个新顶点作为旧边的中点。 对于这样一个EC(G)将不能满足的图,有2n + 1个变量和10n + 2子句。这给了我们无法满足的公式,每个变量的密度接近5个子句。对于子句长度为4的公式,这是一个密度4-SAT集合预期可以满足。 在这个随机模型中,对于n个变量集,每个长度为4的可能子句,构造一个公式,让它成为公式的一部分,概率为p。期望密度为5的随机4-SAT公式也很容易求解。
What can we say about the smallest number of variables involved in a nontrivial conflict, allowing us to learn a new clause, in a formula of this kind? We can get a simple lower bound for this by observing that we must at least set all variables corresponding to the edges of some cycle C in G before a nontrivial conflict can arise. Getting more accurate lower bounds is harder, but this tells us that the girth of the graph can be used to control the size of conflict sets. 在一个非微不足道的冲突中,我们可以用最小的变量数量来学习一个新的子句,在这种公式中,我们能说什么呢? 我们可以得到一个简单的下界,通过观察,我们必须至少设置对应于G中某个循环C的边的所有变量,才能产生非平凡的冲突。 获得更精确的下界是比较困难的,但这告诉我们,图的周长可以用来控制冲突集的大小。
This immediately draws our attention to 4-regular graphs, keeping the clause length low, of high girth. If the girth of G is g we will at first not be able to learn any nontrivial clauses of length less than, at least, g. If the SAT-solver we are using has a cut–off length for the clauses it learns, i.e. it does no retain any conflict clauses with more than some number k of literals, it will in fact be prevented from learning any new clauses at all, thus reducing it to a pure DPLL procedure. 这立即将我们的注意力吸引到4规则图上,保持子句长度低,周长高。如果G的周长是G,我们首先将无法学习任何长度至少小于G的非平凡子句。如果我们正在使用的sat -求解器对它学习的子句有一个截断长度,即它不保留任何冲突子句,其字面量不超过k个,那么它实际上将无法学习任何新的子句,从而将其简化为一个纯粹的DPLL过程。
If the solver uses restarts and removes long clauses when it performs a restart it will in a similar way risk changing into a DPLL procedure with learning, but with an effective run time corresponding to the time between consecutive restarts. 如果求解器在执行重新启动时使用重新启动并删除长子句,那么它将以类似的方式冒着变成具有学习功能的DPLL过程的风险,但其有效运行时间对应于连续重新启动之间的时间间隔。
With this in mind it seems natural to use formulae of the form EC(G) based on 4-regular graphs, as described, as challenging unsatisfiable instances for SAT-solvers. If the formulae are to be tuned for hardness then using graphs with high girth, or at least few short cycles, also seems natural. If hard solvable instances are desired one could apply the methods of [14] on EC(G). The paper [14] presents a construction which, based on a hard unsatisfiable formula builds a solvable formula which is hard for a broad class of solvers. 考虑到这一点,使用基于4正则图的EC(G)形式的公式似乎是很自然的,正如所描述的,作为sat求解者的不满意的挑战实例。如果要调整公式的硬度,那么使用高周长的图,或者至少有几个短周期的图,似乎也很自然。如果需要难解的实例,可以采用的方法 在EC (G)[14]。论文[14]给出了一个构造,它在一个难解不满足公式的基础上,建立了一个对广义求解器来说难解的可解公式。
|
|
|
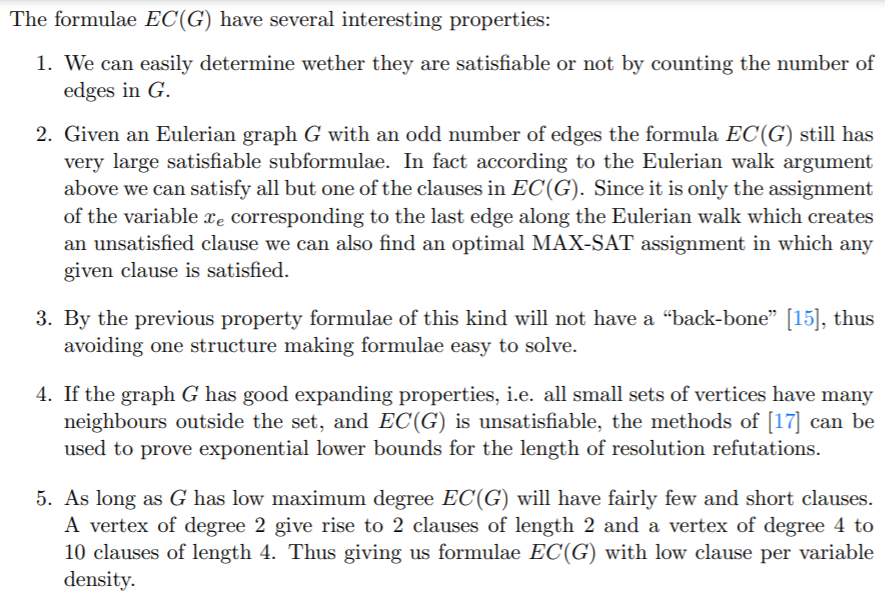
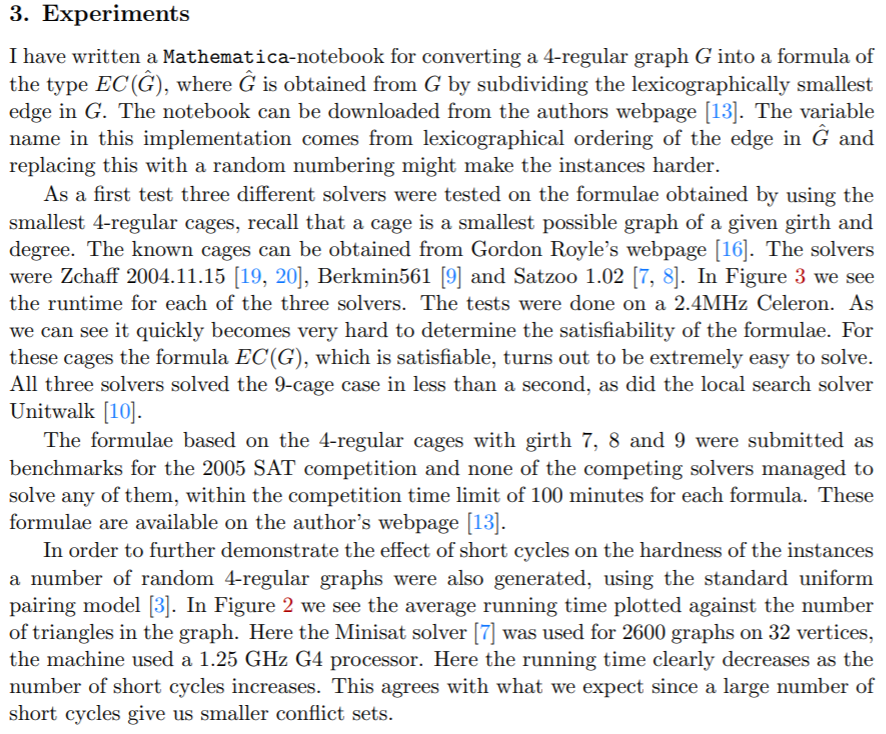
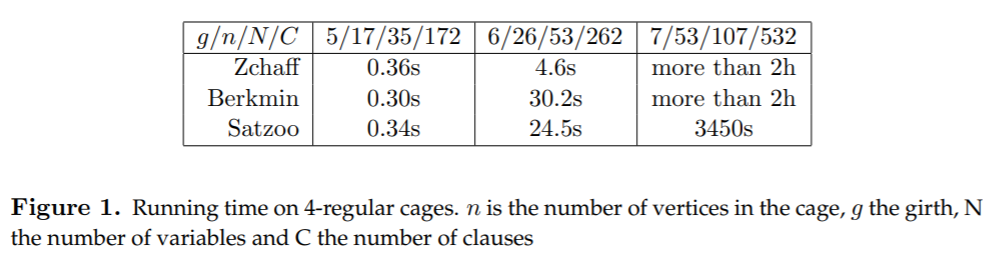
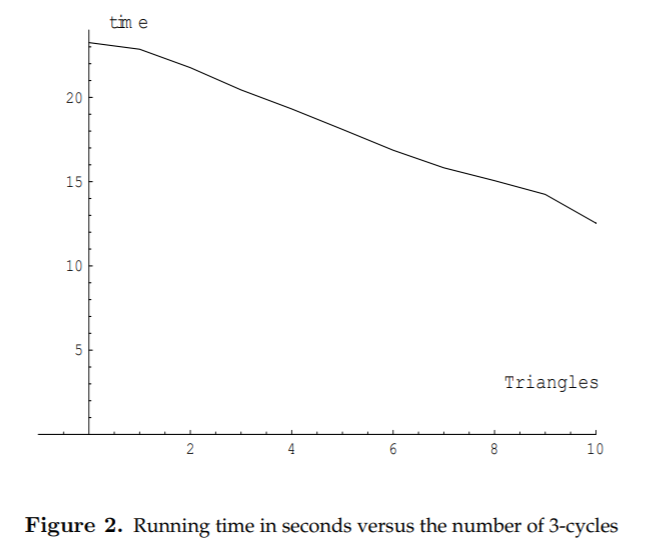
3. Experiments
|
|
|
4. Conclusions and further directions Both our discussion in 2.1 and the experiments in the previous section clearly indicate the importance of small dense structures, such as triangles in the Eulerian graphs, for the hardness of an instance of the type considered here. However we would expect this to be true in greater generality. Let us make this a bit more precise. Given a k-SAT formula F let us look at its clause-variable incidence graph, i.e., the graph with one vertex for each variable, one vertex for each clause, and an edge between a variable-vertex and a clause-vertex if the variable is present in the clause. Given a subset S of the variables we define F[S], the induced subformula of F on S, to be the set of clauses C in F such that C only contains variables present in S. The density of the formula F[S] is as usual the number of clauses in F[S] divided by |S|. If a formula F has many dense subformulae a clause learning DPLL-solver has a good chance of learning new short conflict clauses due to conflicts within a dense local part. However for a given clause length k there exist a density r(k) such that a formula with clauses of length at most k and density less than r(k) is always satisfiable. So, if an unsatisfiable formula F lacks such local dense parts a solver will initially have to assign values to many variables in order to find a conflict and learn a new clause. In this case the learnt clauses will also tend to be quite long. If we look at random k-SAT instances of fixed clause to variable density we will only expect a small number of short cycles in the clause-variable incidence graph, actually an asymptotically size independent number, just as in the case of random regular graphs [3]. As a consequence, the density of F[S] will typically be very small, often less than 1, for variables sets S of small size. Likewise our formulae EC(G) will lack small dense subformulae when the underlying graph G has high girth. If we instead use random regular graphs G we know, [3], that there will typically exist only a small number of cycles in G and no small subgraphs denser than a cycle. In this case the formulae EC(G) will thereby inherit expected properties much like those for random k-SAT. Thus some of the experimentally observed hardness of random unsatisfiable instances may well come from their lack of local dense subformulae. Here an experimental study of running times for unsolvable random instances versus the number of short cycles in their incidence graphs could be interesting. In this context, it is interesting to note that for satisfiable instances this lack of local dense parts is considered to be responsible for some of the success of randomized algorithms like the survey propagation method [4]. An interesting possibility would be to make more explicit use of the structure of the clause-variable incidence graph of an instance in resolution methods as well. This could e.g. be used to control the choice of resolution variable in the DPLL procedure. In order to reduced the risk of the kind of trap the EC(G)-formulae represent for clause learning SAT-solvers some simple modifications can be added. One addition which seems cost effective would be to keep track of a running mean of the size of the clauses discovered during the learning process and keep all clauses not much larger than the current mean. That way long clauses would still be kept for formulae in which no short clauses are possible to learn without first learning long clauses. However, the general problem faced here is still the fundamental limitations of solvers which do not use a proof system stronger than general resolution. While producing more optimised resolution based solvers is undoubtedly a worthwhile undertaking it is becoming more and more important to find efficient algorithms based on stronger proof systems.
References [1] Dimitris Achlioptas and Yuval Peres. The threshold for random k-SAT is 2 k log 2−O(k). J. Amer. Math. Soc., 17(4):947–973 (electronic), 2004. [2] Paul Beame, Henry Kautz, and Ashish Sabharwal. Towards understanding and harnessing the potential of clause learning. J. Artificial Intelligence Res., 22:319–351 (electronic), 2004. [3] B´ela Bollob´as. Random graphs, volume 73 of Cambridge Studies in Advanced Mathematics. Cambridge University Press, Cambridge, second edition, 2001. [4] A. Braunstein, M. M´ezard, and R. Zecchina. Survey propagation: an algorithm for satisfiability. Random Structures Algorithms, 27(2):201–226, 2005. [5] Martin Davis, George Logemann, and Donald Loveland. A machine program for theorem-proving. Comm. ACM, 5:394–397, 1962. [6] Martin Davis and Hilary Putnam. A computing procedure for quantification theory. J. Assoc. Comput. Mach., 7:201–215, 1960. [7] Nicklas Een and Niklas S¨orensson. Webpage for satzoo and minisat at chalmers university. http://www.cs.chalmers.se/∼een/Satzoo/. [8] Nicklas Een and Niklas S¨orensson. An extensible sat-solver. In Lecture Notes in Computer Science, 2919,SAT 2003, pages 502–518, 2004. [9] Eugene Goldberg and Yakov Novikov. Webpage for berkmin. http://heigold.tripod.com/BerkMin.html. [10] Edward A. Hirsch and Arist Kojevnikov. UnitWalk: a new SAT solver that uses local search guided by unit clause elimination. Ann. Math. Artif. Intell., 43(1-4):91–111, 2005. Theory and applications of satisfiability testing. [11] Hans Kleine Buning ¨ and Theodor Lettman. Propositional logic: deduction and algorithms, volume 48 of Cambridge Tracts in Theoretical Computer Science. Cambridge University Press, Cambridge, 1999. Translated from the 1994 German original by the authors. [12] Anton Kotzig. Moves without forbidden transitions in a graph. Mat. Casopis ˇ Sloven. Akad. Vied, 18:76–80, 1968. [13] Klas Markstr¨om. Web page of current author, see combinatorial data and programs. http://www.math.umu.se/∼klasm. [14] Edward A. Hirsch Michael Alekhnovich and Dmitry Itsykson. Exponential lower bounds for the running time of DPLL algorithms on satisfiable formulas. In International Colloquium on Automata, Languages and Programming,ICALP 2004, pages 84–96, 2004. [15] R´emi Monasson, Riccardo Zecchina, Scott Kirkpatrick, Bart Selman, and Lidror Troyansky. Determining computational complexity from characteristic “phase transitions”. Nature, 400(6740):133–137, 1999. [16] Gordon Royle. Gordon royles homepages for combinatorial data. http://www.cs.uwa.edu.au/∼gordon/remote/cubics/index.html. [17] Alasdair Urquhart. Hard examples for resolution. J. Assoc. Comput. Mach., 34(1):209– 219, 1987. [18] Douglas B. West. Introduction to graph theory. Prentice Hall Inc., Upper Saddle River, NJ, 1996. [19] Lintao Zhang. webpage for zchaff at princeton university. http://www.princeton.edu/∼chaff/zchaff.html. [20] Lintao Zhang, Conor F. Madigan, Matthew W. Moskewicz, and Sharad Malik. Efficient conflict driven learning in boolean satisfiability solver. In International Conference on Computer Aided Design, ICCAD 2001, pages 279–285, 2001. |
|