逻辑回归&线性支持向量机
代码:
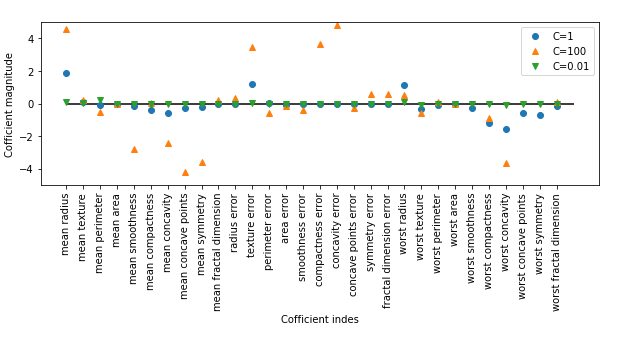
1 # -*- coding: utf-8 -*- 2 """ 3 Created on Tue Jul 17 10:13:20 2018 4 5 @author: zhen 6 """ 7 8 from sklearn.linear_model import LogisticRegression 9 from sklearn.svm import LinearSVC 10 import mglearn 11 import matplotlib.pyplot as plt 12 13 x, y = mglearn.datasets.make_forge() 14 15 fig, axes = plt.subplots(1, 2, figsize=(10,3)) 16 # 线性支持向量机与逻辑回归进行比较 17 for model, ax in zip([LinearSVC(), LogisticRegression()], axes): 18 clf = model.fit(x, y) 19 mglearn.plots.plot_2d_separator(clf, x, fill=False, eps=0.5, ax=ax, alpha=0.7) 20 mglearn.discrete_scatter(x[:, 0], x[:, 1], y, ax=ax) 21 ax.set_title("{}".format(clf.__class__.__name__)) 22 ax.set_xlabel("Feature 0") 23 ax.set_ylabel("Feature 1") 24 axes[0].legend() 25 26 # 27 from sklearn.datasets import load_breast_cancer 28 from sklearn.model_selection import train_test_split 29 cancer = load_breast_cancer() 30 31 x_train, x_test, y_train, y_test = train_test_split(cancer.data, cancer.target, stratify=cancer.target, random_state=42) 32 # 使用默认配置参数 33 log_reg = LogisticRegression().fit(x_train, y_train) 34 35 print("="*25+"逻辑回归(C=1)"+"="*25) 36 print("Training set score:{:.3f}".format(log_reg.score(x_train, y_train))) 37 print("Test set score:{:.3f}".format(log_reg.score(x_test, y_test))) 38 39 # 使用配置参数C=100 40 log_reg_100 = LogisticRegression(C=100).fit(x_train, y_train) 41 42 print("="*25+"逻辑回归(C=100)"+"="*25) 43 print("Training set score:{:.3f}".format(log_reg_100.score(x_train, y_train))) 44 print("Test set score:{:.3f}".format(log_reg_100.score(x_test, y_test))) 45 46 # 使用配置参数C=0.01 47 log_reg_001 = LogisticRegression(C=0.01).fit(x_train, y_train) 48 49 print("="*25+"逻辑回归(C=0.01)"+"="*25) 50 print("Training set score:{:.3f}".format(log_reg_001.score(x_train, y_train))) 51 print("Test set score:{:.3f}".format(log_reg_001.score(x_test, y_test))) 52 print("="*25+"逻辑回归&线性支持向量机"+"="*25) 53 # 可视化 54 fig, axes = plt.subplots(1, 1, figsize=(10,3)) 55 plt.plot(log_reg.coef_.T, 'o', label="C=1") 56 plt.plot(log_reg_100.coef_.T, '^', label="C=100") 57 plt.plot(log_reg_001.coef_.T, 'v', label="C=0.01") 58 plt.xticks(range(cancer.data.shape[1]), cancer.feature_names, rotation=90) 59 plt.hlines(0, 0, cancer.data.shape[1]) 60 61 plt.ylim(-5, 5) 62 63 plt.xlabel("Cofficient indes") 64 plt.ylabel("Cofficient magnitude") 65 66 plt.legend()
结果:




 浙公网安备 33010602011771号
浙公网安备 33010602011771号