以HR,HigherCI,LowerCI为例,表格中要有这三列
先导入表格lq_2
正态化数据分布
lnhr <- log(lq_2[,"HR"])
lnuci <- log(lq_2[,"HigherCI"])
lnlci <- log(lq_2[,"LowerCI"])
selnhr <- (lnuci-lnlci)/(2*1.96)
MetaHR = metagen(TE = lnhr$HR,
seTE = selnhr$HigherCI,
sm = "HR",
data = lq_2,
# studlab = paste(lq_1$Clinic_Pt_Im,lq_1$P.value,sep = " P:"),
studlab = lq_2$`Clinic-Pt-Rad`,
random = T,
#backtransf = T
)
图像风格设置
settings.meta('JAMA')
settings.meta('RevMan5')
settings.meta('meta4')
settings.meta('reset') # 重置风格
绘制森林图(经测试选择某种风格后修改森林图参数会无效)
具体参数可用 ?forest 查询 按照自己的需求调整
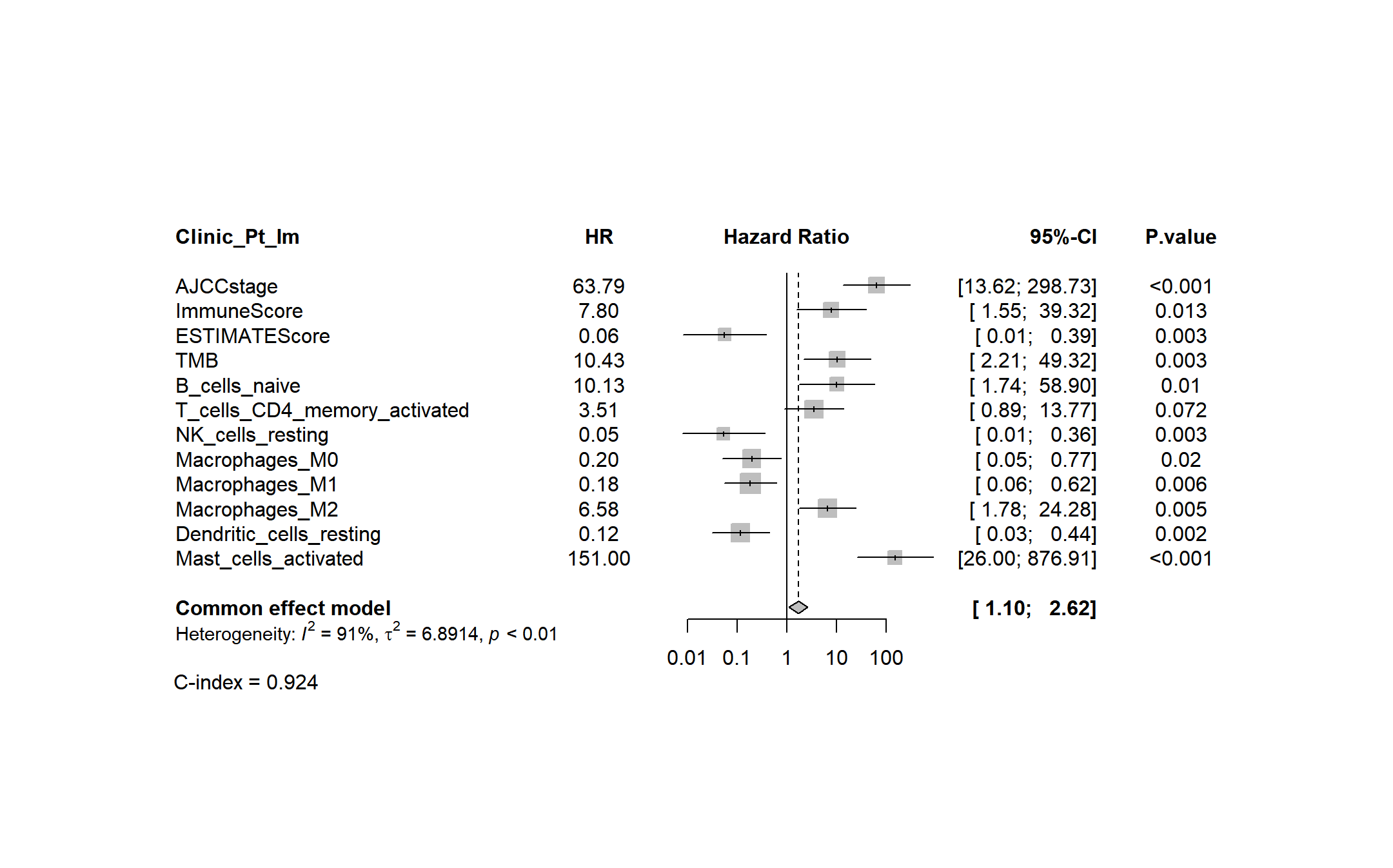
forest(MetaHR,
# xlab = "hahahaha",
label.left = "C-index = 0.840 ",
#label.right = " i am right",
colgap.studlab = "2cm",
#colgap.forest.left = "0.2cm",
colgap.forest.right = "0.5cm",
colgap.right = "1cm",
random = FALSE,
leftlabs = c("Clinic_Pt_Rad", "HR"),
leftcols = c("studlab","HR"),
rightcols = c("ci","P.value"),
)
效果图
参考视频Meta分析自己用-森林图 R语言(高分SCI绘图模仿)03




