python实现决策树
什么是决策树?
决策树是一种基本的分类和回归方法。以分类决策树为例:

决策树通常包含哪三个步骤?
特征选择、决策树的生成和决策树的修剪
决策树与if-then规则?

直接以一个例子看看数如何构建决策树的:
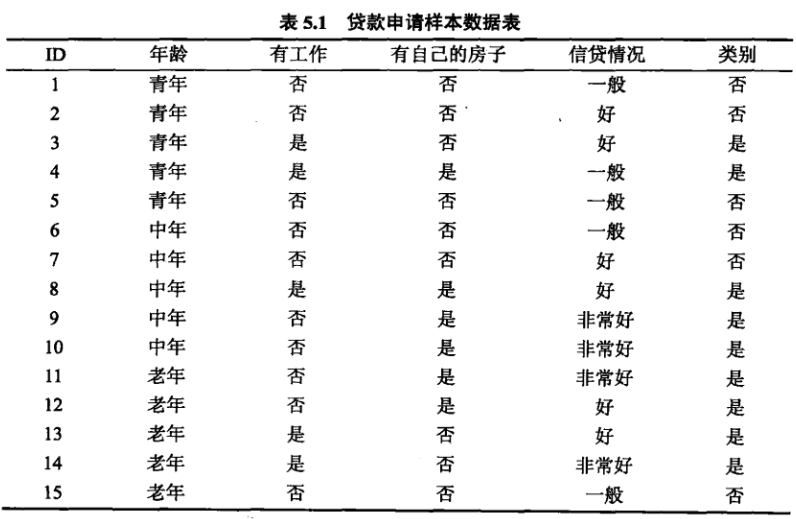
根据不同的特征可以有不同的决策树:
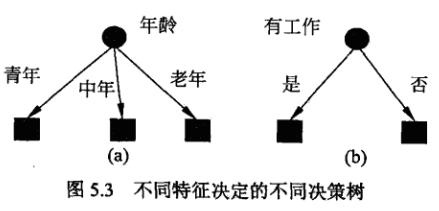
那么如何从根节点开始选择特征进行决策树的构建呢?
最基础的是使用信息增益来表示。
首先得了解熵和条件熵的定义。
熵:用于表示随机变量不确定性的度量 。假设X是一个取值有限的随机变量,其概率分布为:
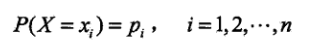
那么随机变量的熵的定义是:


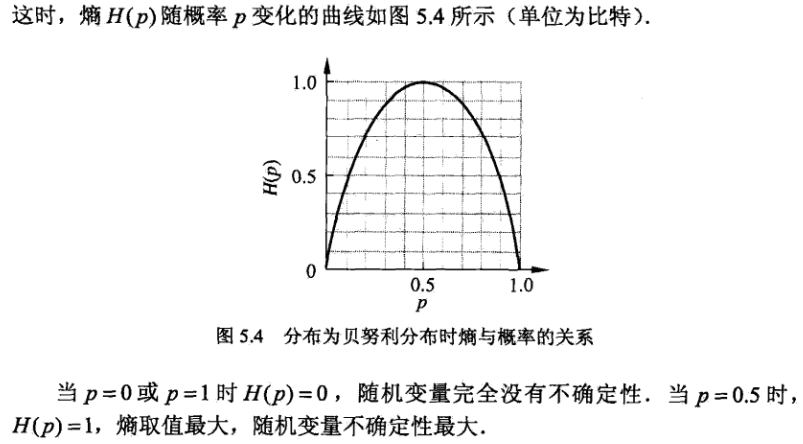

什么是信息增益?

信息增益有什么作用呢?

计算步骤?

这里以上述表格中的数据为例:
我们最终需要的是分为是否会申请贷款,针对于是否需要申请贷款(即经验熵)为:
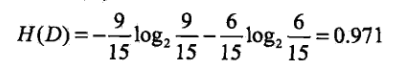
然后我们分别计算每一个特征的条件经验熵(也就是每一个特征对于数据集D的信息增益) ,分别以A1,A2,A3,A4标识年龄、有工作、有自己方法、信贷情况4个特征,则有:



信息增益存在的问题?

那么什么是信息增益比?

提到决策树就需要了解到ID3、C4.5和CART三种。其中ID3就是使用信息增益来进行特征选择,而C4.5使用的是信息增益比进行选择。

ID3生成的决策树如下:

由于ID3只有决策树的生成过程,因此容易过拟合。
CART算法?


以分类为例,CART使用基尼指数来进行特征选择:


还是以上述的数据集进行计算:



还有其剪枝算法,就不列举了。
上述解释摘自:统计学习方法。
下面是代码实现,代码来源: https://github.com/eriklindernoren/ML-From-Scratch
from __future__ import division, print_function import numpy as np from mlfromscratch.utils import divide_on_feature, train_test_split, standardize, mean_squared_error from mlfromscratch.utils import calculate_entropy, accuracy_score, calculate_variance class DecisionNode(): """Class that represents a decision node or leaf in the decision tree Parameters: ----------- feature_i: int Feature index which we want to use as the threshold measure. threshold: float The value that we will compare feature values at feature_i against to determine the prediction. value: float The class prediction if classification tree, or float value if regression tree. true_branch: DecisionNode Next decision node for samples where features value met the threshold. false_branch: DecisionNode Next decision node for samples where features value did not meet the threshold. """ def __init__(self, feature_i=None, threshold=None, value=None, true_branch=None, false_branch=None): self.feature_i = feature_i # Index for the feature that is tested self.threshold = threshold # Threshold value for feature self.value = value # Value if the node is a leaf in the tree self.true_branch = true_branch # 'Left' subtree self.false_branch = false_branch # 'Right' subtree # Super class of RegressionTree and ClassificationTree class DecisionTree(object): """Super class of RegressionTree and ClassificationTree. Parameters: ----------- min_samples_split: int The minimum number of samples needed to make a split when building a tree. min_impurity: float The minimum impurity required to split the tree further. max_depth: int The maximum depth of a tree. loss: function Loss function that is used for Gradient Boosting models to calculate impurity. """ def __init__(self, min_samples_split=2, min_impurity=1e-7, max_depth=float("inf"), loss=None): self.root = None # Root node in dec. tree # Minimum n of samples to justify split self.min_samples_split = min_samples_split # The minimum impurity to justify split self.min_impurity = min_impurity # The maximum depth to grow the tree to self.max_depth = max_depth # Function to calculate impurity (classif.=>info gain, regr=>variance reduct.) self._impurity_calculation = None # Function to determine prediction of y at leaf self._leaf_value_calculation = None # If y is one-hot encoded (multi-dim) or not (one-dim) self.one_dim = None # If Gradient Boost self.loss = loss def fit(self, X, y, loss=None): """ Build decision tree """ self.one_dim = len(np.shape(y)) == 1 self.root = self._build_tree(X, y) self.loss=None def _build_tree(self, X, y, current_depth=0): """ Recursive method which builds out the decision tree and splits X and respective y on the feature of X which (based on impurity) best separates the data""" largest_impurity = 0 best_criteria = None # Feature index and threshold best_sets = None # Subsets of the data # Check if expansion of y is needed if len(np.shape(y)) == 1: y = np.expand_dims(y, axis=1) # Add y as last column of X Xy = np.concatenate((X, y), axis=1) n_samples, n_features = np.shape(X) if n_samples >= self.min_samples_split and current_depth <= self.max_depth: # Calculate the impurity for each feature for feature_i in range(n_features): # All values of feature_i feature_values = np.expand_dims(X[:, feature_i], axis=1) unique_values = np.unique(feature_values) # Iterate through all unique values of feature column i and # calculate the impurity for threshold in unique_values: # Divide X and y depending on if the feature value of X at index feature_i # meets the threshold Xy1, Xy2 = divide_on_feature(Xy, feature_i, threshold) if len(Xy1) > 0 and len(Xy2) > 0: # Select the y-values of the two sets y1 = Xy1[:, n_features:] y2 = Xy2[:, n_features:] # Calculate impurity impurity = self._impurity_calculation(y, y1, y2) # If this threshold resulted in a higher information gain than previously # recorded save the threshold value and the feature # index if impurity > largest_impurity: largest_impurity = impurity best_criteria = {"feature_i": feature_i, "threshold": threshold} best_sets = { "leftX": Xy1[:, :n_features], # X of left subtree "lefty": Xy1[:, n_features:], # y of left subtree "rightX": Xy2[:, :n_features], # X of right subtree "righty": Xy2[:, n_features:] # y of right subtree } if largest_impurity > self.min_impurity: # Build subtrees for the right and left branches true_branch = self._build_tree(best_sets["leftX"], best_sets["lefty"], current_depth + 1) false_branch = self._build_tree(best_sets["rightX"], best_sets["righty"], current_depth + 1) return DecisionNode(feature_i=best_criteria["feature_i"], threshold=best_criteria[ "threshold"], true_branch=true_branch, false_branch=false_branch) # We're at leaf => determine value leaf_value = self._leaf_value_calculation(y) return DecisionNode(value=leaf_value) def predict_value(self, x, tree=None): """ Do a recursive search down the tree and make a prediction of the data sample by the value of the leaf that we end up at """ if tree is None: tree = self.root # If we have a value (i.e we're at a leaf) => return value as the prediction if tree.value is not None: return tree.value # Choose the feature that we will test feature_value = x[tree.feature_i] # Determine if we will follow left or right branch branch = tree.false_branch if isinstance(feature_value, int) or isinstance(feature_value, float): if feature_value >= tree.threshold: branch = tree.true_branch elif feature_value == tree.threshold: branch = tree.true_branch # Test subtree return self.predict_value(x, branch) def predict(self, X): """ Classify samples one by one and return the set of labels """ y_pred = [self.predict_value(sample) for sample in X] return y_pred def print_tree(self, tree=None, indent=" "): """ Recursively print the decision tree """ if not tree: tree = self.root # If we're at leaf => print the label if tree.value is not None: print (tree.value) # Go deeper down the tree else: # Print test print ("%s:%s? " % (tree.feature_i, tree.threshold)) # Print the true scenario print ("%sT->" % (indent), end="") self.print_tree(tree.true_branch, indent + indent) # Print the false scenario print ("%sF->" % (indent), end="") self.print_tree(tree.false_branch, indent + indent) class XGBoostRegressionTree(DecisionTree): """ Regression tree for XGBoost - Reference - http://xgboost.readthedocs.io/en/latest/model.html """ def _split(self, y): """ y contains y_true in left half of the middle column and y_pred in the right half. Split and return the two matrices """ col = int(np.shape(y)[1]/2) y, y_pred = y[:, :col], y[:, col:] return y, y_pred def _gain(self, y, y_pred): nominator = np.power((y * self.loss.gradient(y, y_pred)).sum(), 2) denominator = self.loss.hess(y, y_pred).sum() return 0.5 * (nominator / denominator) def _gain_by_taylor(self, y, y1, y2): # Split y, y_pred = self._split(y) y1, y1_pred = self._split(y1) y2, y2_pred = self._split(y2) true_gain = self._gain(y1, y1_pred) false_gain = self._gain(y2, y2_pred) gain = self._gain(y, y_pred) return true_gain + false_gain - gain def _approximate_update(self, y): # y split into y, y_pred y, y_pred = self._split(y) # Newton's Method gradient = np.sum(y * self.loss.gradient(y, y_pred), axis=0) hessian = np.sum(self.loss.hess(y, y_pred), axis=0) update_approximation = gradient / hessian return update_approximation def fit(self, X, y): self._impurity_calculation = self._gain_by_taylor self._leaf_value_calculation = self._approximate_update super(XGBoostRegressionTree, self).fit(X, y) class RegressionTree(DecisionTree): def _calculate_variance_reduction(self, y, y1, y2): var_tot = calculate_variance(y) var_1 = calculate_variance(y1) var_2 = calculate_variance(y2) frac_1 = len(y1) / len(y) frac_2 = len(y2) / len(y) # Calculate the variance reduction variance_reduction = var_tot - (frac_1 * var_1 + frac_2 * var_2) return sum(variance_reduction) def _mean_of_y(self, y): value = np.mean(y, axis=0) return value if len(value) > 1 else value[0] def fit(self, X, y): self._impurity_calculation = self._calculate_variance_reduction self._leaf_value_calculation = self._mean_of_y super(RegressionTree, self).fit(X, y) class ClassificationTree(DecisionTree): def _calculate_information_gain(self, y, y1, y2): # Calculate information gain p = len(y1) / len(y) entropy = calculate_entropy(y) info_gain = entropy - p * \ calculate_entropy(y1) - (1 - p) * \ calculate_entropy(y2) return info_gain def _majority_vote(self, y): most_common = None max_count = 0 for label in np.unique(y): # Count number of occurences of samples with label count = len(y[y == label]) if count > max_count: most_common = label max_count = count return most_common def fit(self, X, y): self._impurity_calculation = self._calculate_information_gain self._leaf_value_calculation = self._majority_vote super(ClassificationTree, self).fit(X, y)
运行主函数:
from __future__ import division, print_function import numpy as np from sklearn import datasets import matplotlib.pyplot as plt import sys import os import sys sys.path.append("/content/drive/My Drive/learn/ML-From-Scratch/") # Import helper functions from mlfromscratch.utils import train_test_split, standardize, accuracy_score from mlfromscratch.utils import mean_squared_error, calculate_variance, Plot from mlfromscratch.supervised_learning import ClassificationTree def main(): print ("-- Classification Tree --") data = datasets.load_iris() X = data.data y = data.target X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.4) clf = ClassificationTree() clf.fit(X_train, y_train) y_pred = clf.predict(X_test) accuracy = accuracy_score(y_test, y_pred) print ("Accuracy:", accuracy) Plot().plot_in_2d(X_test, y_pred, title="Decision Tree", accuracy=accuracy, legend_labels=data.target_names) if __name__ == "__main__": main()
运行结果:
-- Classification Tree --
Accuracy: 0.9

回归主函数:
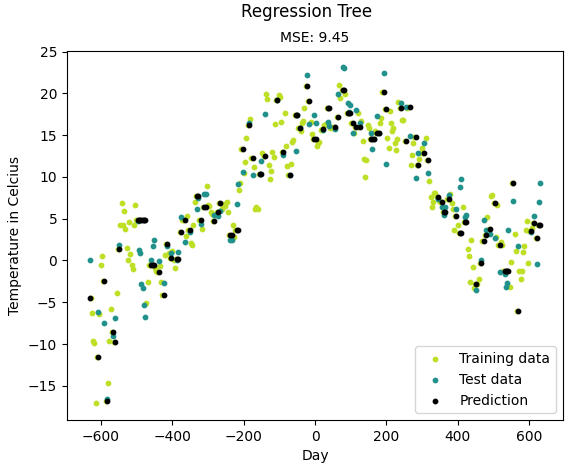
from __future__ import division, print_function import numpy as np import matplotlib.pyplot as plt import pandas as pd import sys sys.path.append("/content/drive/My Drive/learn/ML-From-Scratch/") from mlfromscratch.utils import train_test_split, standardize, accuracy_score from mlfromscratch.utils import mean_squared_error, calculate_variance, Plot from mlfromscratch.supervised_learning import RegressionTree def main(): print ("-- Regression Tree --") # Load temperature data data = pd.read_csv('mlfromscratch/data/TempLinkoping2016.txt', sep="\t") time = np.atleast_2d(data["time"].values).T temp = np.atleast_2d(data["temp"].values).T X = standardize(time) # Time. Fraction of the year [0, 1] y = temp[:, 0] # Temperature. Reduce to one-dim X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3) model = RegressionTree() model.fit(X_train, y_train) y_pred = model.predict(X_test) y_pred_line = model.predict(X) # Color map cmap = plt.get_cmap('viridis') mse = mean_squared_error(y_test, y_pred) print ("Mean Squared Error:", mse) # Plot the results # Plot the results m1 = plt.scatter(366 * X_train, y_train, color=cmap(0.9), s=10) m2 = plt.scatter(366 * X_test, y_test, color=cmap(0.5), s=10) m3 = plt.scatter(366 * X_test, y_pred, color='black', s=10) plt.suptitle("Regression Tree") plt.title("MSE: %.2f" % mse, fontsize=10) plt.xlabel('Day') plt.ylabel('Temperature in Celcius') plt.legend((m1, m2, m3), ("Training data", "Test data", "Prediction"), loc='lower right') plt.savefig("test2.png") plt.show() if __name__ == "__main__": main()
结果:
-- Regression Tree --
Mean Squared Error: 9.445229357798167