7. The Singular Value Decomposition(SVD)
7.1 Singular values and Singular vectors
The SVD separates any matrix into simple pieces.
A is any m by n matrix, square or rectangular, Its rank is r.
Choices from the SVD
\(u_i\)— the left singular vectors (unit eigenvectors of \(AA^T\))
\(v_i\)— the right singular vectors (unit eigenvectors of \(A^TA\))
\(\sigma_i\)— singular values (square roots of the equal eigenvalues of \(AA^T\) and \(A^TA\))
The rank of A is equal to numbers of \(\sigma _i\)
example:
7.2 Bases and Matrices in the SVD
Keys:
-
The SVD produces orthonormal basis of \(u's\) and $ v's$ for the four fundamental subspaces.
- \(u_1,u_2,...,u_r\) is an orthonormal basis of the column space. (\(R^m\))
- \(u_{r+1},...,u_{m}\) is an orthonormal basis of the left nullspace. (\(R^m\))
- \(v_1,v_2,...,v_r\) is an orthonormal basis of the row space. (\(R^n\))
- \(v_{r+1},...,u_{n}\) is an orthonormal basis of the nullspace.(\(R^n\))
-
Using those basis, A can be diagonalized :
Reduced SVD: only with bases for the row space and column space.
\[A = U_r \Sigma_r V_r^T \\ U = \left [ \begin{matrix} u_1&\cdots&u_r\\ \end{matrix}\right] , \Sigma_r = \left [ \begin{matrix} \sigma_1&&\\&\ddots&\\&&\sigma_r \end{matrix}\right], V_r^T=\left [ \begin{matrix} v_1\\ \vdots \\ v_r \end{matrix}\right] \\ \Downarrow \\ A = \left [ \begin{matrix} u_1&\cdots&u_r\\ \end{matrix}\right] \left [ \begin{matrix} \sigma_1&&\\&\ddots&\\&&\sigma_r \end{matrix}\right] \left [ \begin{matrix} v_1\\ \vdots \\ v_r \end{matrix}\right] \\ = u_1\sigma_1v_{1}^T + u_2\sigma_2v_{2}^T + \cdots + u_r\sigma_rv_r^T \]Full SVD: include four subspaces.
\[A = U \Sigma V^T \\ U = \left [ \begin{matrix} u_1&\cdots&u_r&\cdots&u_n\\ \end{matrix}\right] , \Sigma_r = \left [ \begin{matrix} \sigma_1&&\\&\ddots&\\&&\sigma_r \\ &&&\ddots \\ &&&&\sigma_n \end{matrix}\right], V^T=\left [ \begin{matrix} v_1\\ \vdots \\ v_r \\ \vdots \\ v_m \end{matrix}\right] \\ \Downarrow \\ A = \left [ \begin{matrix} u_1&\cdots&u_r&\cdots&u_n\\ \end{matrix}\right] \left [ \begin{matrix} \sigma_1&&\\&\ddots&\\&&\sigma_r \\ &&&\ddots \\ &&&&\sigma_n \end{matrix}\right] \left [ \begin{matrix} v_1\\ \vdots \\ v_r \\ \vdots \\ v_m \end{matrix}\right] \\ = u_1\sigma_1v_{1}^T + u_2\sigma_2v_{2}^T + \cdots + u_r\sigma_rv_r^T\cdots + u_n\sigma_n v_n^{T} + \cdots + u_m\sigma_mv_m^T \]example: \(A=\left [ \begin{matrix} 3&0 \\ 4&5 \end{matrix}\right]\), r=2
\[A^TA =\left [ \begin{matrix} 25&20 \\ 20&25 \end{matrix}\right], AA^T =\left [ \begin{matrix} 9&12 \\ 12&41 \end{matrix}\right] \\ \lambda_1 = 45, \sigma_1 = \sqrt{45}, v_1 = \frac{1}{\sqrt{2}} \left [ \begin{matrix} 1 \\ 1 \end{matrix}\right], u_1 = \frac{1}{\sqrt{10}} \left [ \begin{matrix} 1 \\ 3 \end{matrix}\right]\\ \lambda_2 = 5, \sigma_2 = \sqrt{5} , v_2 = \frac{1}{\sqrt{2}} \left [ \begin{matrix} -1 \\ 1 \end{matrix}\right], u_2 = \frac{1}{\sqrt{10}} \left [ \begin{matrix} -3 \\ 1 \end{matrix}\right]\\ \Downarrow \\ U = \frac{1}{\sqrt{10}} \left [ \begin{matrix} 1&-3 \\ 3&1 \end{matrix}\right], \Sigma = \left [ \begin{matrix} \sqrt{45}& \\ &\sqrt{5} \end{matrix}\right], V = \frac{1}{\sqrt{2}} \left [ \begin{matrix} 1&-1 \\ 1&1 \end{matrix}\right] \]
7.3 The geometry of the SVD
- \(A = U\Sigma V^T\) factors into (rotation)(stretching)(rotation), the geometry shows how A transforms vectors x on a circle to vectors Ax on an ellipse.
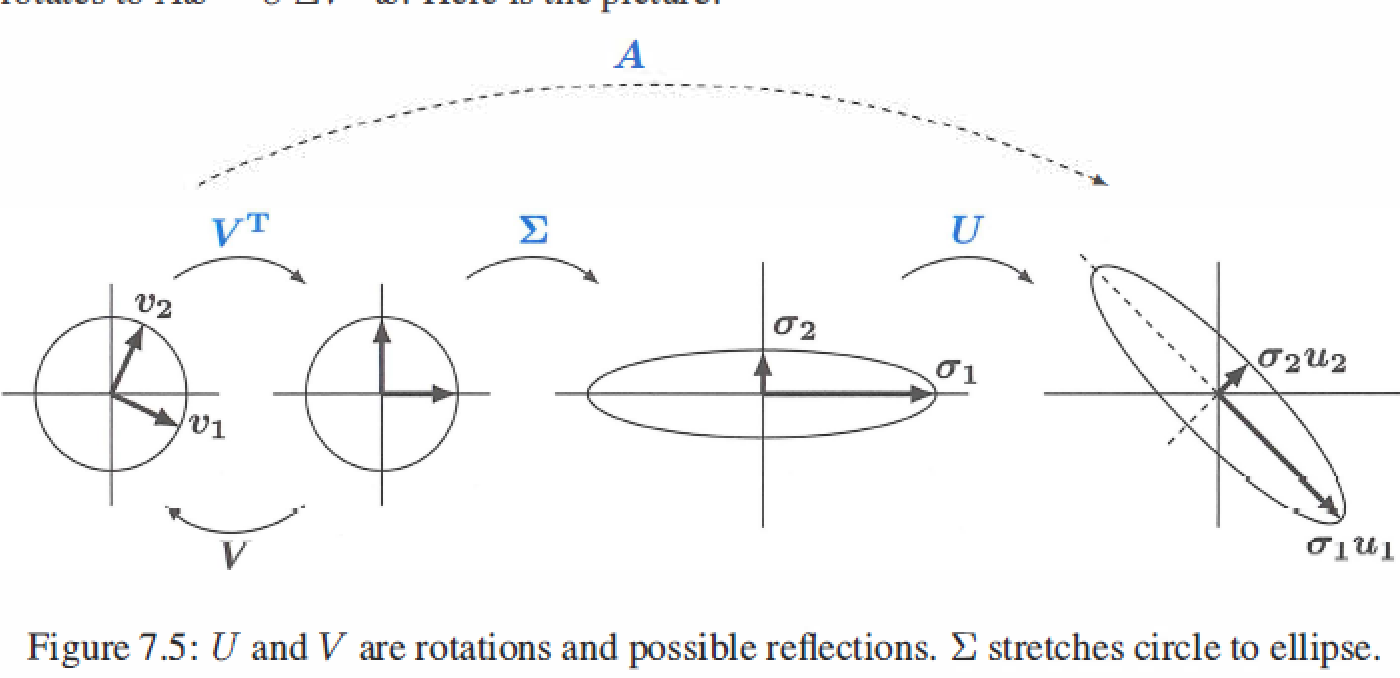
-
Polar decomposition factors A into QS : rotation \(Q=UV^T\) times streching \(S=V \Sigma V^T\).
\[V^TV = I \\ A = U\Sigma V^T = (UV^T)(V\Sigma V^T) = (Q)(S) \]Q is orthogonal and inclues both rotations U and \(V^T\), S is symmetric positive semidefinite and gives the stretching directions.
If A is invertible, S is positive definite.
-
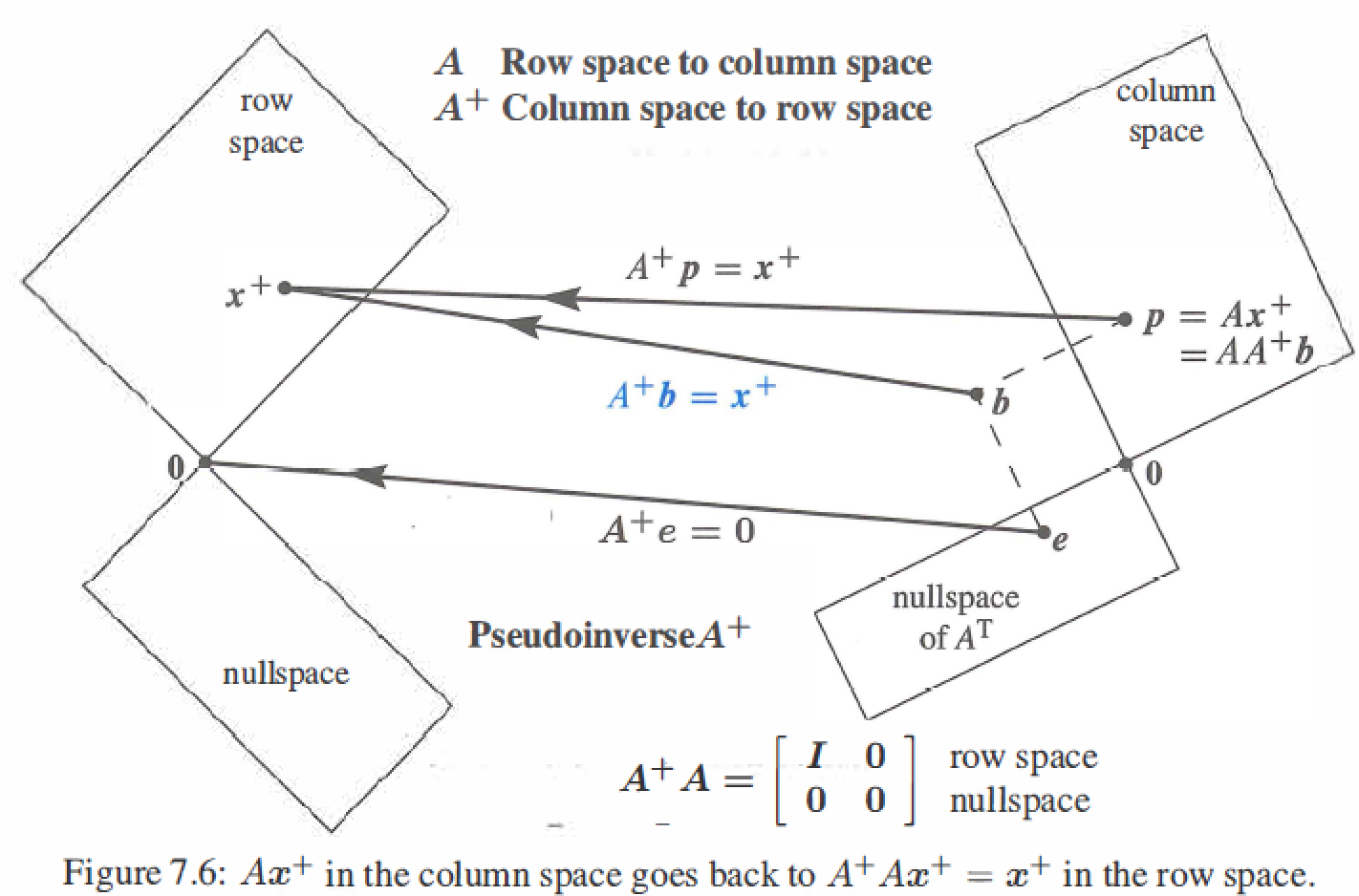
The Pseudoinverse \(A^{+}: AA^{+}=I\)
-
\(Av_i=\sigma_iu_i\) : A multiplies \(v_i\) in the row space of A to give \(\sigma_i u_i\) in the column space of A.
-
If \(A^{-1}\) exists, \(A^{-1}u_i=\frac{v_i}{\sigma}\) : \(A^{-1}\) multiplies \(u_i\) in the row space of \(A^{-1}\) to give \(\sigma_i u_i\) in the column space of \(A^{-1}\), \(1/\sigma_i\) is singular values of \(A^{-1}\).
-
Pseudoinverse of A: if \(A^{-1}\) exists, then \(A^{+}\) is the same as \(A^{-1}\)
\[A^{+} = V \Sigma^{+}U^{T} = \left [ \begin{matrix} v_1&\cdots&v_r&\cdots&v_n\\ \end{matrix}\right] \left [ \begin{matrix} \sigma_1^{-1}&&\\&\ddots&\\&&\sigma_r^{-1} \\ &&&\ddots \\ &&&&\sigma_n^{-1} \end{matrix}\right] \left [ \begin{matrix} u_1\\ \vdots \\ u_r \\ \vdots \\ u_m \end{matrix}\right] \\ \]
-
7.4 Principal Component Analysis ( PCA by the SVD)
PCA gives a way to understand a data plot in dimension m, applications mostly are human genetics \ face recognition\ finance \ model order reduction (computation) .
The sample covariance matrix \(S=AA^T/(n-1)\)
The crucial connection to linear algebra is in the singular values and singular vectors of A, which comes from the eigenvalues \(\lambda=\sigma^2\) and the eigenvectors u of the sample covariance matrix \(S=AA^T/(n-1)\)
-
The total variance in the data is the sum of all eigenvalues and of sample variances \(s^2\) :
\[T = \sigma_1^2 + \cdots + \sigma_m^2 = s_1^2 + \cdots + s_m^2 = trace(diagonal \ \ sum) \] -
The first eigenvector \(u_1\) of S points in the most significant direction of the data.That direction accounts for a fraction \(\sigma_1^2/T\) of the total variance.
-
The next eigenvectors \(u_2\) (orthogonal to \(u_1\)) accounts for a small fraction \(\sigma_2^2/T\).
-
Stop when those fractions are small. You have the R directions that explain most of the data.The n data points are very near an R-dimensional subspace with basis \(u_1, \cdots, u_R\), which are the principal components.
-
R is the "effective rank" of A. The true rank r is probably m or n : full rank matrix.
example: \(A = \left[ \begin{matrix} 3&-4&7&-1&-4&-3 \\ 7&-6&8&-1&-1&-7 \end{matrix} \right]\) has sample covariance \(S=AA^T/5 = \left [ \begin{matrix} 20&25 \\ 25&40 \end{matrix}\right]\)
The eigenvalues of S are 57 and 3,so the first rank one piece \(\sqrt{57}u_1v_1^T\) is much larger than the second piece \(\sqrt{3}u_2v_2^T\).
The leading eigenvector \(u_1 = (0.6,0.8)\) shows the direction that you see in the scatter graph.
The SVD of A (centered data) shows the dominant direction in the scatter plot.
The second eigenvector \(u_2\) is perpendicular to \(u_1\). The second singular value \(\sigma_2=\sqrt{3}\) measures the spread across the dominant line.








