Evolutionary approaches towards AI: past, present, and future
Evolutionary approaches towards AI: past, present, and future
2019-10-06 07:28:13
This blog is from: https://towardsdatascience.com/evolutionary-approaches-towards-ai-past-present-and-future-b23ccb424e98
Table of Contents
- Introduction
- Genetics and natural selection
- Evolutionary computation
- — Evolution strategies
- — Genetic algorithms with direct encoding
- — Genetic algorithms with indirect encoding
- — Open-endedness (here’s where it gets really interesting!)
- — What’s still missing?
- Conclusion
- References
Introduction
Since roughly 2012 [1], the explosive growth in AI has been almost entirely driven by neural network (deep learning) models trained by back-propagation (“backprop”). This includes models for image classification, automatic speech recognition, language translation, robotics, and autonomous agents that can play single or multiplayer games.
Increasingly, however, models are constructed using methods based on aspects of evolution, as observed in biology. Such approaches predate the deep learning era but have only recently been scaled up to the point where they are competitive with backprop-trained deep learning models.
In this blog post we discuss some of these evolutionary approaches, compare and contrast them to biological evolution and organism development, and speculate how they may ultimately bring about AI models with even greater (or additional) capabilities and energy-efficiencies than traditional deep learning models.
Genetics and natural selection
At its simplest, Darwin’s theory states that evolution is driven by small variations in traits that are amplified by natural selection. Organisms with beneficial traits are more likely to reproduce and thus pass on those traits to offspring than are organisms that have detrimental traits.
Darwin was unaware of how traits were passed from parents to offspring (making his insights all the more impressive), but we now know that an organism’s genotype, along with its developmental environment, is responsible for its phenotype (physical and behavioral traits). In general, new genotypes arise in offspring via random mutation of parent DNA, a mixing of genes from multiple parents (sexual reproduction), or both.
Evolutionary computation
Out of scientists’ understanding of biological evolution come computational optimization models inspired by evolution. Among the simplest of these are evolution strategies. Greater and more diverse modeling complexity is found in genetic algorithms. In both cases, the goal is to optimize a fitness function intended to measure the performance of artificial organisms on some specific task. An alternative approach is to forgo fitness functions and instead utilize rich, open environments with multiple agents of one or more species in which agents compete solely to survive and reproduce.
Evolution strategies
Evolution strategies are a class of optimization algorithms in which, at every iteration (generation), a population of parameter vectors (genotypes) is perturbed (mutated) and their fitness function value is evaluated [2, 3]. The highest scoring parameter vectors are then recombined to form the population for the next generation and this procedure is iterated until the objective is fully optimized. In Covariance-Matrix Adaptation Evolution Strategy (CMA-ES) the distribution of model parameters is represented by a covariance matrix. In each generation, the model parameters of individual agents are drawn from the distribution. After determination of which agents have the highest fitness scores, the covariance matrix is updated based on the parameter values of those best agents. For a great 2-D visualization, see this terrific Otoro blog post.
Despite the simplicity of this approach, it can sometime be competitive with relatively modern, large-scale, deep learning models trained with reinforcement learning methods, as shown by OpenAI [4] and described in this post. The evolution strategies approach has some nice properties relative to the reinforcement learning methods in that evolution strategies are easy to implement and scale across cores/CPUs, models train quickly, and the method does not utilize gradients (training on tasks with discrete outputs is difficult when using gradient-based methods).

Above are a few videos of 3D humanoid walkers trained with evolution strategies by OpenAI (videos taken from their blog post). Results have quite a bit of variety, based on which local minimum the optimization ends up converging into. As will be noted later in this post, however, none of movements look very natural in contrast to those of biological animals, some of which begin fluid walking and running within minutes or hours after birth.
Genetic algorithms with direct encoding
While the term genetic algorithm may be used in different manners by different researchers and practitioners, we use it in a very general manner herein. In each generation the algorithm: (1) selects for the top N agents from a population of size P (N < P) based on the fitness function, (2) produces a new generation of agents by reproducing from those top agents individually (asexually) or in pairs (sexually), and (3) during reproduction, offspring genes are varied by mutation, crossover (mixing genes of two partner parents), or both.
Another distinction between evolution strategies and genetic algorithms is that in evolution strategies, the genomes of the population are represented by a probability distribution. Typically this means that all members of a population in a given generation will be found in a single cluster within the parameter (genome) space. In contrast, there is no such constraint on the population under the genetic algorithm. In practice, however, a single population cluster typically evolves unless the environment or additional algorithmic components promote(s) population diversity (in which case, multiple “species” may emerge).
In many applications of genetic algorithms, the genetype-to-phenotype is direct, meaning that each gene directly codes for one parameter of the agent’s model. In fact, a gene and its phenotypic expression may be identical, e.g., a numeric value that represents a weight or bias term in a deep learning model. The models trained using evolution strategies, as described in the section above, use direct encoding.
In contrast, biology is based on indirect encoding. For example, genes composed of DNA do not directly code for the strength of synapses between neurons in the brain. Rather, they code for proteins which together function to implement the development of the brain (and its synapses) as well as learning rules by which synapses strengthen or weaken based on the organism’s experiences. We’ll come back to examples of indirect encoding for AI in the following section.
In 2017, Ken Stanley and Jeff Clune — long time advocates of “neuroevolutionary” methods for evolving neural network parameters — demonstrated the potential for genetic algorithms with direct encoding to perform well on numerous Atari games, including games that were difficult to solve by reinforcement learning (Q-learning or policy gradients) [5]. Their team at Uber AI Labs used a simple genetic algorithm in which genetic mutations were introduced into asexually-produced offspring by adding Gaussian noise to the parameters of the parent network.
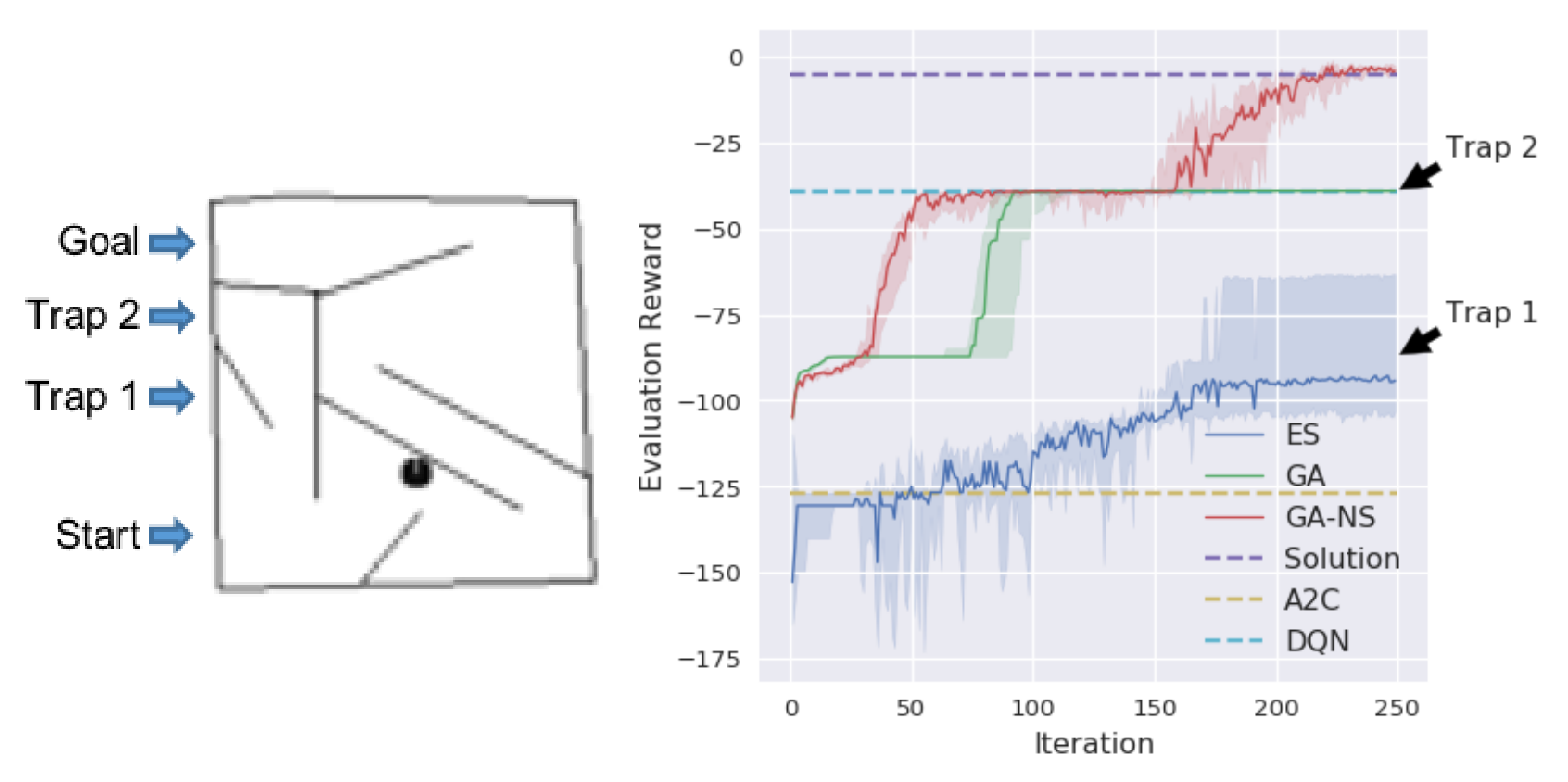
In addition to evolving agents to play Atari games, the team from Uber AI Labs evolved agents to complete a relatively simple maze, albeit one with two different “traps.” Agents trained with an evolution strategy (ES) consistently get stuck by Trap 1 and never evolve further. Agents trained with a genetic algorithm (GA) do better, but get stuck in Trap 2. When agents are chosen for reproduction based not only on their fitness score but also their display of novel behaviors (GA-NS), the species ultimately evolves the ability to complete the maze. Agents trained with two different reinforcement learning methods (A2C and DQN) do not learn to complete the maze.
The Uber team additionally examined the effect of rewarding agents (allowing them to reproduce) for exhibiting novel behaviors. They demonstrated that despite the fact that such agents often scored poorly on the traditional fitness function, this approach benefited the overall population over time (over generations). Genetic algorithms that assign value for behavioral novelty are a type of quality-diversity algorithm [6], and such algorithms are an active area of study. The notion is that maintaining a diverse repertoire of behaviors across the population provides a pool from which new, more complex behaviors may emerge that could benefit an offspring organism, despite little or even negative value of the more simplistic behaviors upon which the complex behavior is built. [Note that this is largely a heuristic approach, as it is not clear what natural forces would promote the retention of “worthless” behavioral phenotypes across generations.]
As an aside, the Uber team uses a creative and efficient method of retaining the genotype of thousands of agents of large size (many millions of neural network parameters per agent). Namely, they maintain a record of the random number generator seeds used to create the initial generation of agents, and the seeds to create each set of mutations. Thus, an individual agent can be recreated from a vector of seeds, and the agent can be stored as such, rather than storing the agent’s model parameters directly.
Genetic algorithms with indirect encoding
Generally speaking, genetic algorithms that utilize indirect encoding models have not been as successful at direct encoding models (on modern, large-scale problems). Nonetheless indirect encoding models may prove to be very powerful because they have the potential to compactly encode for complex, sophisticated models. For example, while a set of genes within an organism may be fixed, the protein products of genes may interact across time and space (in the body of the organism) in a combinatorial fashion, allowing nearly endless possibilities. We highlight two examples of indirect encoding below.
HyperNEAT
The evolutionary computation methods discussed so far have a genome of fixed size. That is, a neural network of fixed architecture. The genes define the parameters of this fixed architecture but do not define any aspect of the architecture and thus the genetic algorithm has not way of growing, shrinking, or modifying that architecture. In 2002, Stanley and Miikkulainen introduced the NeuroEvolution of Augmenting Topologies (NEAT) [7]. NEAT defines a mapping between genes and the connections within an neural network, and can accommodate the evolutionary addition of new genes that define new connections or nodes.
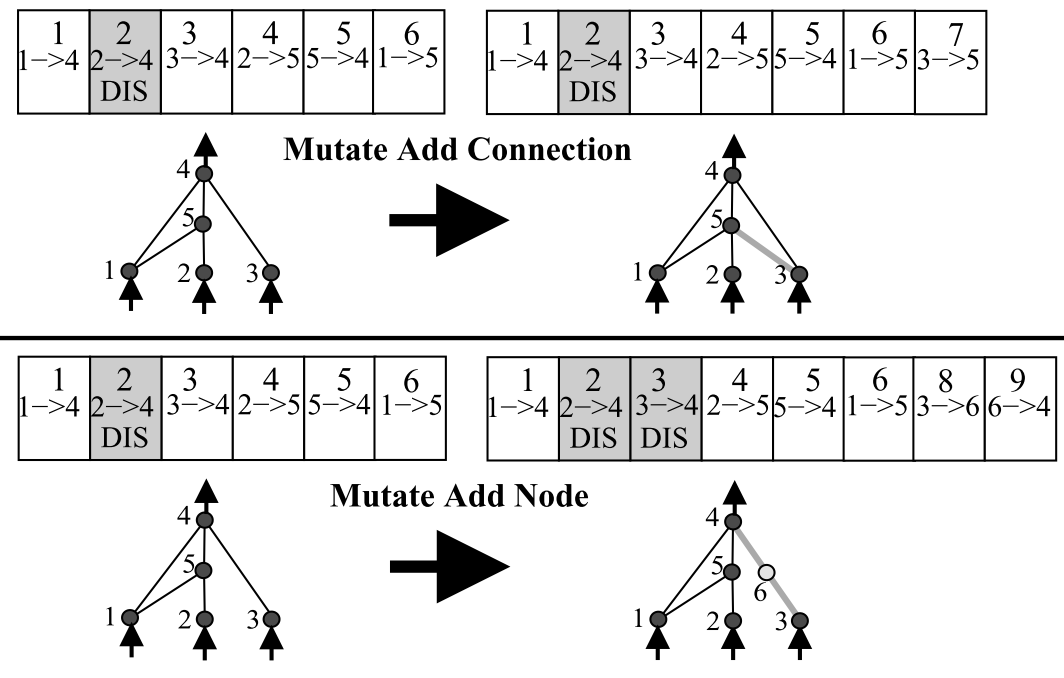
NEAT defines rules that map genes to a network architecture in addition to values of network weight and bias terms. Under a genetic algorithm, the network can grow by adding connections and nodes.
However, NEAT is a direct encoding model, with each gene defining the connection weight between two nodes. While some genes can take on a “disabled” state, this can be viewed as encoding a weight value of zero. This takes us to HyperNEAT.
Under HyperNEAT [8], the output of a NEAT-trained network defines the weights of a secondary network. This secondary network is the one used to perform the desired task. In the simplest version, this secondary “task” network has nodes arranged in a two-dimensional space such that each node can be identified by its (x, y) coordinates. The first network, dubbed a compositional pattern-producing network (CPPN), takes four inputs that define the locations of two nodes (i and j) in the task network: (xi, yi) and (xj, yj). The output of the CPPN is the value of the weight that connects these two points. Thus, NEAT can be used to evolve a CPPN that directs the “development” of the task network. The task network, not the CPPN, is evaluated by the fitness function.
With the HyperNEAT approach, a relatively small CPPN network can define a complex task network of arbitrary scale in density. As a demonstration of the evolvability of the CPPNs, models have been evolved to produce intricate 2-D images. And while performance of evolved task networks lags behind that of more recent models, HyperNEAT has been used to train models to play Atari games [9] (at roughly the same time that DeepMind demonstrated such models trained by reinforcement learning [10]).
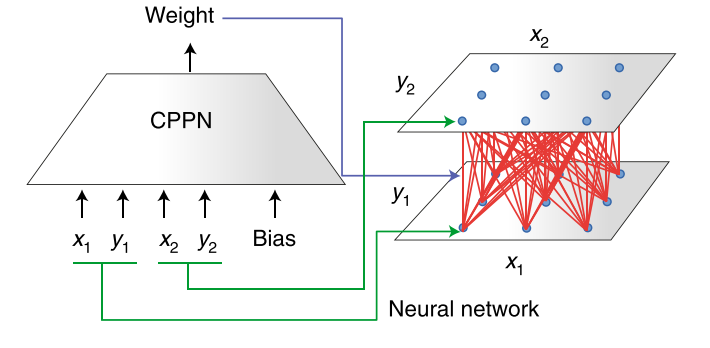
In this HyperNEAT example, the task network (right) is a two-layered network in which neurons connect between the lower and upper layer, but not within layers. The CPPN network (left) defines connection strengths of the task layer for a given pair of task network nodes. The CPPN is a neural network defined by the NEAT genome-to-network mapping, and can be evolved using a genetic algorithm, with a fitness function applied to the task network defined by a given CPPN instantiation.
Organism development
Biological evolution does not establish the phenotypes of mature (adult) organisms directly. Rather, genes indirectly establish an organisms phenotype through development (e.g., prenatal, infancy, adolescence). Genes also play a role during adulthood in response to an organism’s environment (e.g., creating more red blood cells in in an organisms that moves from low to high altitude). While NEAT evolves a population of neural networks, those networks are not actually “grown” from a genome. Rather, they are instantiated directly since NEAT networks are directly encoded.
Researchers such as Jordan Pollack and his former post-doc, Sylvain Cussat-Blanc, are exploring evolutionary computation methods that incorporate organism developmental periods. In a 2015 study [11], they evolved models of gene regulatory networks (GRNs) using a genetic algorithm related to NEAT. Generally, GRNs are networks in which genes (and non-gene DNA, RNA, and proteins) control the expression (translation to proteins) of other genes. Different genes are expressed during different development periods and under different environmental conditions. Thus, what is evolved is not the organisms per se, but the manner in which they are developed. The authors demonstrate superiority over standard genetic algorithms. However, the computational complexity is high and unlikely to scale to more challenging, modern AI tasks using today’s standard hardware.
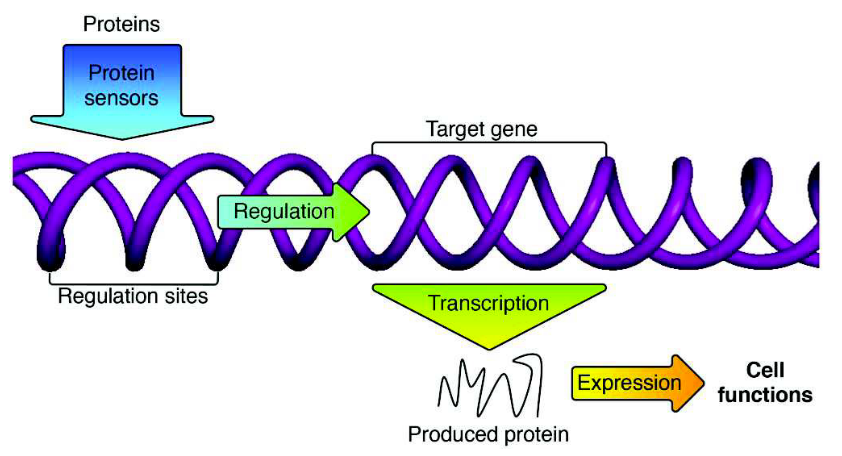
Rather then evolve a genome that directly encoded a neural network (such as NEAT), Pollack and colleagues [11] evolved a gene regulatory network that controlled the development of a neural network, much like biological genomes direct human growth from embryo to adult.
More recently, Miller et al. [12] constructed developmental models for neurons (somas and their dendrites) within neural networks. The developmental models were represented by computer programs that could be evolved using genetic programming. Genetic programming is distinctly different from genetic algorithms and we leave it to the reader to explore this topic elsewhere. Nonetheless, Miller and colleagues took an evolutionary computation approach toward creating development models that construct useful neural networks. The result was networks that could grow new dendrites and neural connections in order to learn new tasks.
Open-endedness
Up to this point we’ve discussed evolutionary approaches that utilize a fitness function to explicitly score the performance of individual agents and determine which will produce offspring for the next generation. Clearly nature has no explicit fitness function. (It does, however, have a sort of implicit inverse fitness function — if organism X produces more offspring than organism Y, then organism X is likely to have higher fitness than organism Y.) That is, evolution in the natural world has produced remarkably intelligent species (humans), as well as species with unique and fascinating physical and innate behavioral traits, with no external guidance.
This observation has motivated the research fields of artificial life and open-ended evolution. Artificial life refers to the study of artificial organisms in real or artificial environments that closely mirror some or many aspects of natural environments. Researchers use this approach to study life as we know it, but also life as it might be otherwise. Open-ended evolution studies often incorporate artificial organisms like those in artificial life studies, and observe the evolution of these organisms under conditions like those that produced biological evolution — namely, reproduction based on direct actions within the environment (finding mates, obtaining enough food to survive and reproduce, evading predators, etc.) rather than based on explicit fitness measures. The field of open-ended evolution is relatively small compared to that of deep learning but is actually a notably older field, with pioneering researchers such as Charles Ofria, Jordan Pollack, Risto Miikkulainen, and their students (several of whom are now leaders in the field) having spent decades in the trenches. For a more complete description of the history and prospects of open-endedness, see this post from Lehman, Stanley, and Soros, and this paper by Jeff Clune [13].
Can artificial organisms evolved in open-ended environments lead to advancements in AI models (agents)? We believe the answer is “yes,” but is conditioned on at least two factors: (1) co-evolution of species and (2) evolution within a rich, diverse, and dynamic environment. (For a different perspective on the matter, see Lisa Soros’s PhD thesis [14].)
Addressing the first factor, environments must house a diversity of agents of different species with distinct needs and capabilities such that co-evolution of species might lead to collaboration within species — a likely prerequisite for the evolution of human-level intelligence. While co-evolution has led to faster and faster cheetahs and impalas, it has also led to intelligent social behavior in wolves, as is exhibited by coordinated hunting in packs.
Due in part to co-evolution, wolves have evolved a social intelligence that allows them to hunt in packs — thereby killing animals much larger than themselves and earning a food reward that benefits the entire pack, not just individual wolves.
Indeed, a very recent study by OpenAI demonstrates the emergence of complex interactive behaviors among agents trained by reinforcement learning, despite a seemingly impoverished reward system. Notably, rewards were given for team (akin to species) performance, not individual performance — much like all the wolves in a pack are rewarded when a large kill is made.
Given the simple task of playing hide-and-seek, these agents learn to collaborate to perform complex task that at first blush, seem to have little to do with the primary hide-and-seek objective. Competition between teams is a driving factor in the emergence of these collaborative within-team behaviors. Note that these agents were trained by reinforcement learning rather than evolutionary computation. However, the relevant point is that competition between teams of agents (or species) can promote novel and complex behaviors.
There is an important contrast between reinforcement learning and evolutionary computation that should be mentioned here. In nature, reinforcement learning serves as a model for how animals learn during their lifetime. When an animal’s behavior is rewarded (via food, shelter, mating, etc.) it is more likely to repeat the behavior, hopefully to its benefit. However, many animal capabilities are acquired through evolution and bestowed during prenatal development or quickly after birth. For example, humans are born with nearly innate sense of “objectness” (though not “object permanence”) — babies do not need to learn that “pixels” (photons on the retina) in close spatial proximity are more likely be part of the same object. An extreme example of innateness is the rapidity with which some animals gain complex motor control after birth (within minutes). The bottom line is that biological organism are pre-wired for many abilities. Deep learning models are generally trained from scratch (“tabula rasa”) and narrowly targeted toward a specific application. Constructing AI agents with more general, real-world capabilities might be achieved by first evolving agents to have baseline innate knowledge (physics, emotions, fundamental desires, etc.) at “birth.” The agents could subsequently learn to perform niche tasks through reinforcement learning mechanisms (which, ideally, will have been evolved into the agent — e.g., meta-learning) during their “lifetimes.”
Baby wildebeests gain agile muscle control and navigation abilities with minutes of birth. Clearly evolution has given them an innate sense of gravity, physics, objectness, and an advanced sensorimotor control system. They do no need to learn these concepts and capabilities through their reinforcement learning system.
As previously stated, the second factor we believe to be critical for evolving intelligent agents is a rich, diverse, and dynamic environment. Such an environment contains niche conditions across time and space that certain genetic mutations may prove advantageous within, but would be washed out of the species over subsequent generations if the environment was homogeneous. In turn, the mutated, advantageous phenotypes may also prove to be advantageous in a different niche environment, which organisms could come across due to self-driven movement or due to changes in their local environment (e.g., changes in climate, as in the real world). This is somewhat akin to the previously discussed quality-diversity approaches in genetic algorithms.
What’s still missing?
Beyond what has already been stated, we list here a few aspects of agents, agent environments, and genetic algorithms that we speculate could promote the evolution of agents with capabilities beyond those of current AI models. Some of the listed agent aspects may evolve naturally if the conditions support this, but they could also be imposed directly to accelerate evolution toward the end goal of general artificial intelligence.
Agents and environments
- Parenting and long-development periods: Forcing reproducing agents to care for helpless offspring during a developmental period may promote social interactions, communication (language), collaborations (between parents of a given offspring, between parents and children, and even between non-related parents and children — it takes a village to raise a child), etc.
- Individual recognition: Agents should be able to identify other individual agents of the same species (via “visual” or other features that are unique to each agent, and are derived from an agent’s genome). If agents are able to distinguish one another, then they can relate an individual agent to that agent’s behavior, potentially allowing for the evolution of trust, collaboration, care-giving, etc. (but also distrust, trickery, and collusion).
- Communications medium: The environment should allow some modality by which agents could evolve a means of communication (a language). This could be acoustical, visual, or even tactile in nature. This may be necessary for agents to evolve complex social interactions and collaborations.
Genes and genetic algorithms
- Indirect encoding with combinatorial genes: The use of genes that interact to produce a higher-level product (e.g., proteins, or control over production of proteins) may allow for a compact but highly expressive genomes that can be more effectively evolved than a gene model in which each gene maps to one and only one model parameter or phenotypic trait. In addition, this combinatorial aspect may limit the likelihood of an offspring being completely non-viable (and thus a waste of simulation/evaluation time) due to gene mutation or crossover.
- Genes that instruct development: Related to the above, genes that encode organism development rather than a final, adult organism may be more compact and evolvable (as in HyperNEAT). If the development is also driven by interactions between genes and the environment, this could provide additional selective pressure on genes and thus promote evolution.
- Genomes that can evolve in size: Behaviorally complex organisms may require more genes to define their phenotype (or their development) than those with simplistic behavior. However, an evolution procedure that starts with a simple species and small genome might evolve into a complexly behaving, large-genome species more quickly than a species in which the genome is large at the start of the evolution procedure. Selective pressure for individual genes in a simple species with a large genome may be weak, due to the minimal impact any one gene might have on an organism’s phenotype, thus slowing the evolution.
- Structured selection of reproducing and dying agents: In algorithms that utilized a fitness function, the typical approach is to select the top performing agents for reproduction and kill the remaining agents within that generation. However, under this approach, newly mutated genes that provide a fitness advantage to an agent may still be lost by genetic drift across the species. Recent work [15] suggests that strategically selecting which agent to kill when another agent reproduces (rules of which are represented by a structured “evolutionary graph”) can promote the “amplification” of the new, advantageous, genes across the species and reduce the probability that the beneficial gene will be lost. In rich, open-ended environments, such structure is presumably imposed indirectly by the environment.
Conclusion
Deep learning has inarguably brought about tremendous AI advances in recent years, and more are likely to come. Nonetheless, we believe that evolutionary computation approaches to AI are historically understudied and will ultimately yield similar leaps in AI capabilities — either building on those provided by deep learning, or offering altogether new ones.
Finally, we speculate that evolutionary computation methods could lead to AI that is highly computationally efficient. By evolving agents on a given hardware platform and properly designing the genes-to-instructions mapping, genes that accelerate task completion could be selected for, optimizing performance on that platform.
References
- Krizhevsky, A., Sutskever, I., and Hinton, G. E. ImageNet classification with deep convolutional neural networks. In NIPS, pp. 1106–1114, 2012.
- I. Rechenberg and M. Eigen. Evolutionsstrategie: Optimierung Technischer Systeme nach Prinzipiender Biologischen Evolution. Frommann-Holzboog Stuttgart, 1973.
- H.-P. Schwefel.Numerische optimierung von computer-modellen mittels der evolutionsstrategie.1977.
- Salimans T., Ho J., Chen X., and Sutskever I. Evolution strategies as a scalable alternative to reinforcement learning. arXiv preprint arXiv:1703.03864, 2017.
- Felipe Petroski Such, Vashisht Madhavan, Edoardo Conti, Joel Lehman, Kenneth O Stanley, and Jeff Clune. Deep neuroevolution: Genetic algorithms are a competitive alternative for training deep neural networks for reinforcement learning. arXiv preprint arXiv:1712.06567, 2017.
- Justin K Pugh, Lisa B Soros, and Kenneth O Stanley. Quality diversity: A new frontier for evolutionary computation. Frontiers in Robotics and AI, 3:40, 2016.
- Stanley, K. O. & Miikkulainen, R. Evolving neural networks through augmenting topologies. Evol. Comput. 10, 99–127 (2002).
- Stanley, Kenneth O.; D’Ambrosio, David B.; Gauci, Jason (2009–01–14). “A Hypercube-Based Encoding for Evolving Large-Scale Neural Networks”. Artificial Life. 15 (2): 185–212.
- Hausknecht, M., Lehman, J., Miikkulainen, R. & Stone, P. A neuroevolution approach to general atari game playing. IEEE Trans. Comput. Intell. AI Games 6, 355–366 (2014).
- Mnih, V. et al. Human-level control through deep reinforcement learning. Nature 518, 529–533 (2015).
- Cussat-Blanc, S., Harrington, K. & Pollack, J. Gene regulatory network evolution through augmenting topologies. IEEE Trans. Evolut. Comput. 19, 823–837 (2015).
- Miller, J.F., Wilson, D.G., Cussat-Blanc, S.: Evolving developmental programs that build neural networks for solving multiple problems. In: Banzhaf, W., Spector, L., Sheneman L. (eds.) Genetic Programming Theory and Practice XVI, Chap. TBC. Springer (2019).
- Jeff Clune. AI-GAs: AI-generating algorithms, an alternate paradigm for producing general artificial intelligence. arXiv preprint arXiv:1905.10985, 2019.
- Soros, Lisa, “Necessary Conditions for Open-Ended Evolution” (2018). Electronic Theses and Dissertations. 5965. https://stars.library.ucf.edu/etd/5965.
- Pavlogiannis A, Tkadlec J, Chatterjee K, Nowak MA. Construction of arbitrarily strong amplifiers of natural selection using evolutionary graph theory. Communications Biology. 2018;1(1):71.



 浙公网安备 33010602011771号
浙公网安备 33010602011771号