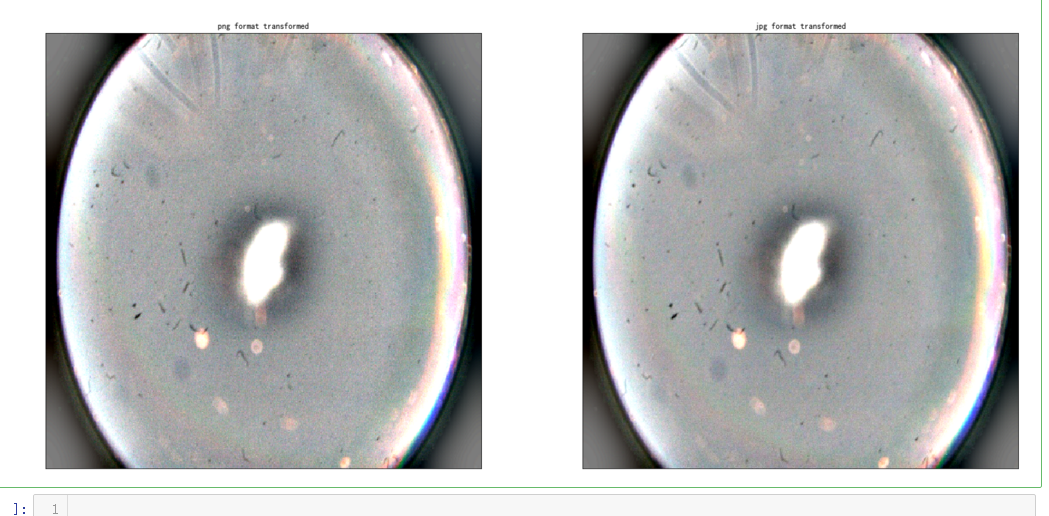
dpi = 80 #inch path_jpg=f"F:\\kaggleDataSet\\diabeticRetinopathy\\resized_train_cropped\\18017_left.jpeg" # too many vessels? path_png=f"F:\\kaggleDataSet\\diabeticRetinopathy\\rescaled_train_896\\18017_left.png" # details are lost image = cv2.imread(path_png) image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB) image = cv2.resize(image, (IMG_SIZE, IMG_SIZE)) image2 = cv2.imread(path_jpg) image2 = cv2.cvtColor(image2, cv2.COLOR_BGR2RGB) image2 = cv2.resize(image2, (IMG_SIZE, IMG_SIZE)) height, width = IMG_SIZE, IMG_SIZE print(height, width) SCALE=1/4 figsize = (width / float(dpi))/SCALE, (height / float(dpi))/SCALE fig = plt.figure(figsize=figsize) ax = fig.add_subplot(2, 2, 1, xticks=[], yticks=[]) ax.set_title('png format original' ) plt.imshow(image, cmap='gray') ax = fig.add_subplot(2, 2, 2, xticks=[], yticks=[]) ax.set_title('jpg format original' ) plt.imshow(image2, cmap='gray') image = load_ben_color(path_png,sigmaX=30) image2 = load_ben_color(path_jpg,sigmaX=30) ax = fig.add_subplot(2, 2, 3, xticks=[], yticks=[]) ax.set_title('png format transformed' ) plt.imshow(image, cmap='gray') ax = fig.add_subplot(2, 2, 4, xticks=[], yticks=[]) ax.set_title('jpg format transformed' ) plt.imshow(image2, cmap='gray')

import json import math import os import cv2 from PIL import Image import numpy as np from keras import layers from keras.applications import DenseNet121 from keras.callbacks import Callback, ModelCheckpoint from keras.preprocessing.image import ImageDataGenerator from keras.models import Sequential from keras.optimizers import Adam import matplotlib.pyplot as plt import pandas as pd from sklearn.model_selection import train_test_split from sklearn.metrics import cohen_kappa_score, accuracy_score import scipy from tqdm import tqdm %matplotlib inline
train_df = pd.read_csv('F:\\kaggleDataSet\\diabeticRetinopathy\\trainLabels19.csv') test_df = pd.read_csv('F:\\kaggleDataSet\\diabeticRetinopathy\\testImages19.csv') print(train_df.shape) print(test_df.shape) test_df.head()
def get_pad_width(im, new_shape, is_rgb=True): pad_diff = new_shape - im.shape[0], new_shape - im.shape[1] t, b = math.floor(pad_diff[0]/2), math.ceil(pad_diff[0]/2) l, r = math.floor(pad_diff[1]/2), math.ceil(pad_diff[1]/2) if is_rgb: pad_width = ((t,b), (l,r), (0, 0)) else: pad_width = ((t,b), (l,r)) return pad_width def preprocess_image(image_path, desired_size=224): im = Image.open(image_path) im = im.resize((desired_size, )*2, resample=Image.LANCZOS) return im
N = test_df.shape[0] x_test = np.empty((N, 224, 224, 3), dtype=np.uint8) for i, image_id in enumerate(tqdm(test_df['id_code'])): x_test[i, :, :, :] = preprocess_image("F:\\kaggleDataSet\\diabeticRetinopathy\\resized test 19\\"+str(image_id)+".jpg")
# model.summary() def load_image_ben_orig(path,resize=True,crop=False,norm255=True,keras=False): image = cv2.imread(path) image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB) image=cv2.addWeighted( image,4, cv2.GaussianBlur( image , (0,0) , 10) ,-4 ,128) if norm255: return image/255 elif keras: #see https://github.com/keras-team/keras-applications/blob/master/keras_applications/imagenet_utils.py for mode #see https://github.com/keras-team/keras-applications/blob/master/keras_applications/xception.py for inception,xception mode #the use of tf based preprocessing (- and / by 127 respectively) will results in [-1,1] so it will not visualize correctly (directly) image = np.expand_dims(image, axis=0) return preprocess_input(image)[0] else: return image.astype(np.int16) return image def transform_image_ben(img,resize=True,crop=False,norm255=True,keras=False): image=cv2.addWeighted( img,4, cv2.GaussianBlur( img , (0,0) , 10) ,-4 ,128) if norm255: return image/255 elif keras: image = np.expand_dims(image, axis=0) return preprocess_input(image)[0] else: return image.astype(np.int16) return image
def display_samples(df, columns=5, rows=2, Ben=True): fig=plt.figure(figsize=(5*columns, 4*rows)) for i in range(columns*rows): image_path = df.loc[i,'id_code'] path = f"F:\\kaggleDataSet\\diabeticRetinopathy\\resized test 19\\"+str(image_path)+".jpg" if Ben: img = load_image_ben_orig(path) else: img = cv2.imread(path) img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB) fig.add_subplot(rows, columns, i+1) plt.imshow(img) plt.tight_layout() display_samples(test_df, Ben=False) display_samples(test_df, Ben=True)




