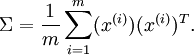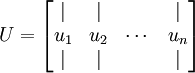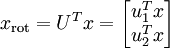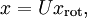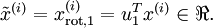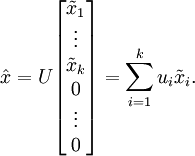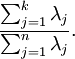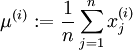PCA
http://deeplearning.stanford.edu/wiki/index.php/PCA
Principal Components Analysis (PCA) is a dimensionality reduction algorithm that can be used to significantly speed up your unsupervised feature learning algorithm.
example
Suppose you are training your algorithm on images. Then the input will be somewhat redundant, because the values of adjacent pixels in an image are highly correlated. Concretely, suppose we are training on 16x16 grayscale image patches. Then
are 256 dimensional vectors, with one feature
corresponding to the intensity of each pixel. Because of the correlation between adjacent pixels, PCA will allow us to approximate the input with a much lower dimensional one, while incurring very little error.
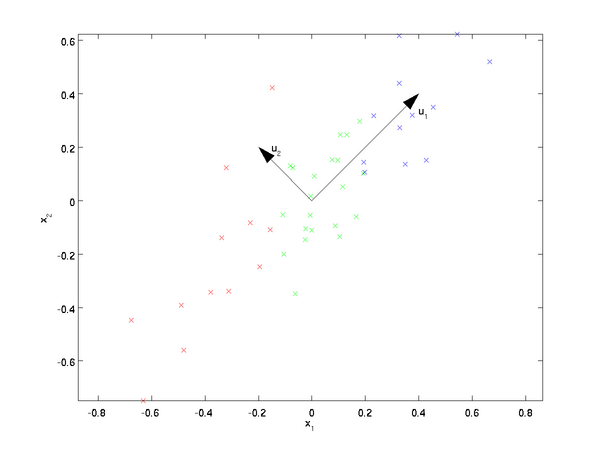
PCA will find a lower-dimensional subspace onto which to project our data. From visually examining the data, it appears that
is the principal direction of variation of the data, and
the secondary direction of variation:
the data varies much more in the direction
than
.
To more formally find the directions 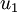 and
and 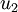 , we first compute the matrix
, we first compute the matrix 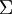 as follows:
as follows:
If 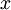 has zero mean, then
has zero mean, then 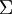 is exactly the covariance matrix of
is exactly the covariance matrix of 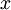 . (The symbol "
. (The symbol "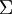 ", pronounced "Sigma", is the standard notation for denoting the covariance matrix. Unfortunately it looks just like the summation symbol, as in
", pronounced "Sigma", is the standard notation for denoting the covariance matrix. Unfortunately it looks just like the summation symbol, as in 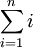 ; but these are two different things.)
; but these are two different things.)
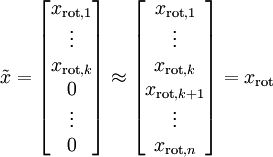
let us compute the eigenvectors of 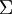 , and stack the eigenvectors in columns to form the matrix
, and stack the eigenvectors in columns to form the matrix 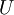 :
:
Here, 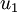 is the principal eigenvector (corresponding to the largest eigenvalue),
is the principal eigenvector (corresponding to the largest eigenvalue), 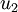 is the second eigenvector, and so on. Also, let
is the second eigenvector, and so on. Also, let 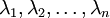 be the corresponding eigenvalues.
be the corresponding eigenvalues.
The vectors 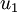 and
and 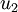 in our example form a new basis in which we can represent the data. Concretely, let
in our example form a new basis in which we can represent the data. Concretely, let 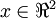 be some training example. Then
be some training example. Then 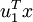 is the length (magnitude) of the projection of
is the length (magnitude) of the projection of 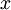 onto the vector
onto the vector 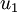 .
.
Similarly, 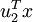 is the magnitude of
is the magnitude of 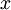 projected onto the vector
projected onto the vector 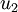 .
.
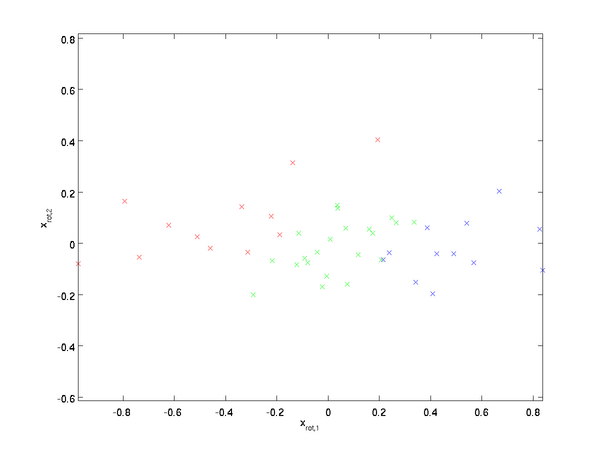
Rotating the Data
Thus, we can represent
in the
-basis by computing
(The subscript "rot" comes from the observation that this corresponds to a rotation (and possibly reflection) of the original data.) Lets take the entire training set, and compute
for every
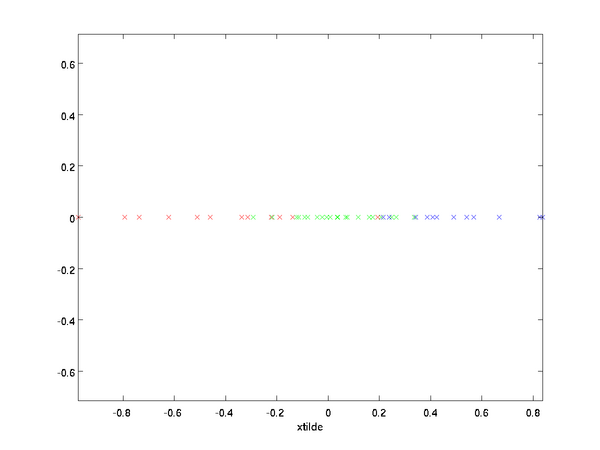
. Plotting this transformed data
, we get:
This is the training set rotated into the
,
basis. In the general case,
will be the training set rotated into the basis
,
, ...,
.
One of the properties of
is that it is an "orthogonal" matrix, which means that it satisfies
. So if you ever need to go from the rotated vectors
back to the original data
, you can compute
because
.
Reducing the Data Dimension
We see that the principal direction of variation of the data is the first dimension
of this rotated data. Thus, if we want to reduce this data to one dimension, we can set
More generally, if
and we want to reduce it to a
dimensional representation
(where
), we would take the first
components of
, which correspond to the top
directions of variation.
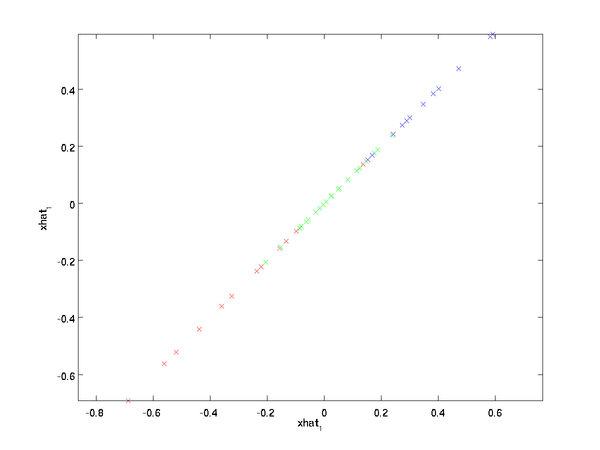
In our example, this gives us the following plot of
(using
):
However, since the final
components of
as defined above would always be zero, there is no need to keep these zeros around, and so we define
as a
-dimensional vector with just the first
(non-zero) components.
This also explains why we wanted to express our data in the
basis: Deciding which components to keep becomes just keeping the top
components. When we do this, we also say that we are "retaining the top
PCA (or principal) components."
Recovering an Approximation of the Data
we can think of
as an approximation to
, where we have set the last
components to zeros. Thus, given
, we can pad it out with
zeros to get our approximation to
. Finally, we pre-multiply by
to get our approximation to
. Concretely, we get
We are thus using a 1 dimensional approximation to the original dataset.
Number of components to retain
To decide how to set
, we will usually look at the percentage of variance retained for different values of
. Concretely, if
, then we have an exact approximation to the data, and we say that 100% of the variance is retained. I.e., all of the variation of the original data is retained. Conversely, if
, then we are approximating all the data with the zero vector, and thus 0% of the variance is retained.
More generally, let
be the eigenvalues of
(sorted in decreasing order), so that
is the eigenvalue corresponding to the eigenvector
. Then if we retain
principal components, the percentage of variance retained is given by:
In our simple 2D example above,
, and
. Thus, by keeping only
principal components, we retained
, or 91.3% of the variance.
PCA on Images
For PCA to work, usually we want each of the features
to have a similar range of values to the others (and to have a mean close to zero). If you've used PCA on other applications before, you may therefore have separately pre-processed each feature to have zero mean and unit variance, by separately estimating the mean and variance of each feature
. However, this isn't the pre-processing that we will apply to most types of images. Specifically, suppose we are training our algorithm on natural images, so that
is the value of pixel
. By "natural images," we informally mean the type of image that a typical animal or person might see over their lifetime.
In detail, in order for PCA to work well, informally we require that (i) The features have approximately zero mean, and (ii) The different features have similar variances to each other. With natural images, (ii) is already satisfied even without variance normalization, and so we won't perform any variance normalization.
Concretely, if
are the (grayscale) intensity values of a 16x16 image patch (
), we might normalize the intensity of each image
as follows:
, for all