R EnhancedVolcano 绘制火山图
火山图是用于差异表达分析结果可视化的一种有效方法。今天,我们来介绍一个用于增强火山图绘制的强大 R 包:EnhancedVolcano ,该包拥有强大的绘图功能,用户可以简单的通过设置颜色、形状、大小和阴影等参数定义不同的绘图属性,此外通过可以通过添加连线的方式有效避免数据点之间的重叠现象。使用 EnhancedVocalno 包绘制的火山图基本可以直接用于文献发表,可以说非常简单又实用的一款神器了。
1. 下载与安装
R 版本:3.6.1。从 Bioconductor 中下载包:
if (!requireNamespace('BiocManager', quietly = TRUE))
install.packages('BiocManager')
BiocManager::install('EnhancedVolcano')
2. 简单使用
2.1 输入数据格式
首先,我们先来介绍一下 EnhancedVolcan o 输入数据格式。EnhancedVolcano 包可以使用多种差异算法(例如 DESeq2 等)的结果作为输入,数据中需包含 log2FC 、Pvalue 或(和) qvalue 结果,示例数据如下:
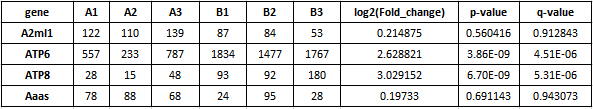
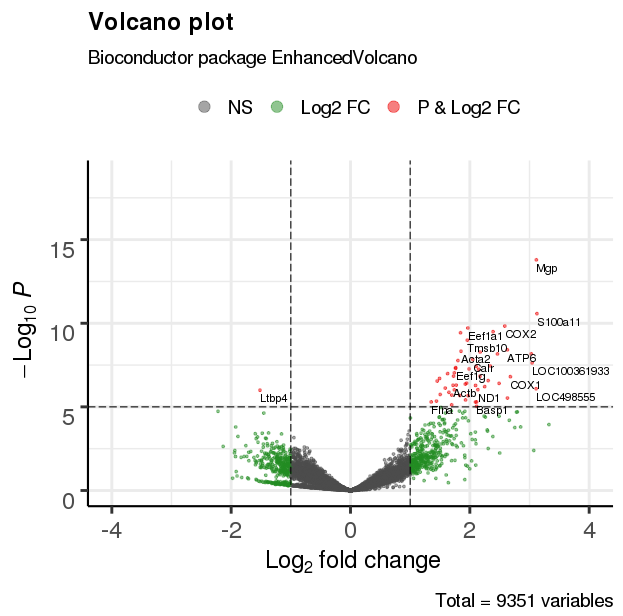
2.2 基础绘图
library(EnhancedVolcano)
res <- read.table(diffexpress,
sep="\t",
head=T,
row.names=1,
check.names=F,
quote="")
EnhancedVolcano(res,
lab = rownames(res),
x = 'log2(Fold_change)',
y = 'p-value',
xlim = c(-4, 4))
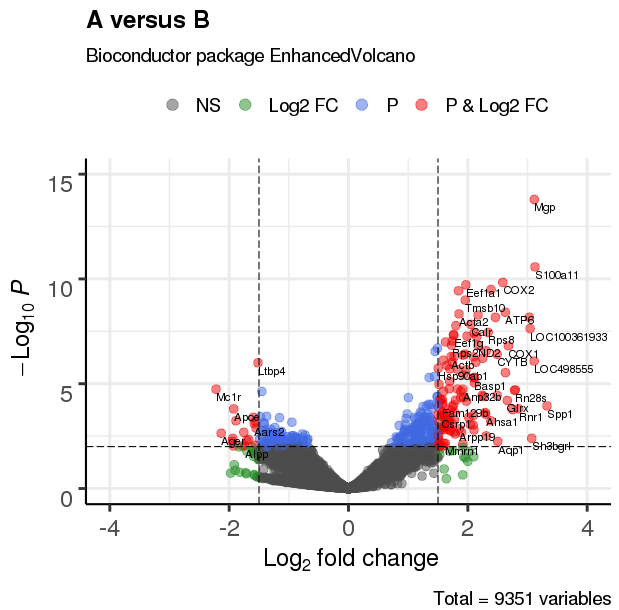
3、进阶功能
3.1 调整阈值,设置点及标签大小
EnhancedVolcano(res,
lab = rownames(res),
x = 'log2(Fold_change)',
y = 'p-value',
xlim = c(-4, 4),
ylim = c(0,15),
title = 'A versus B',
pCutoff = 10e-3,
FCcutoff = 1.5,
transcriptPointSize = 3.0,
transcriptLabSize = 3.0)
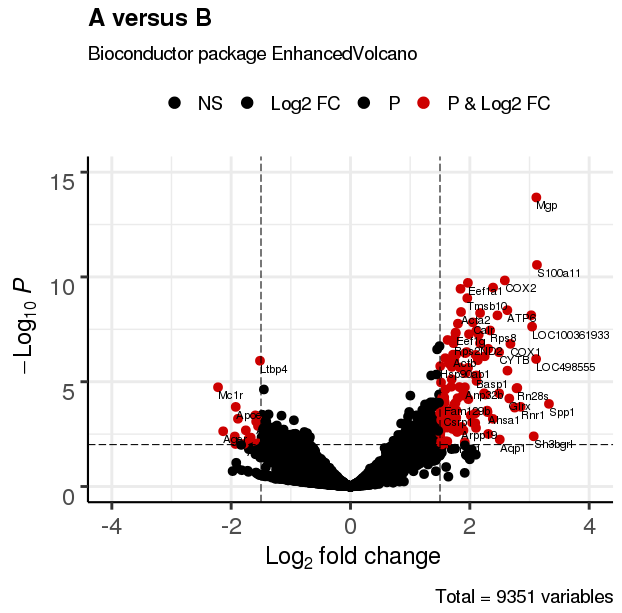
3.2 调整颜色及点透明度
EnhancedVolcano(res,
lab = rownames(res),
x = 'log2(Fold_change)',
y = 'p-value',
xlim = c(-4, 4),
ylim = c(0,15),
title = 'A versus B',
pCutoff = 10e-3,
FCcutoff = 1.5,
transcriptPointSize = 3.0,
transcriptLabSize = 3.0,
col = c('black', 'black', 'black', 'red3'),
colAlpha = 1)
3.3 调整绘图点形状
EnhancedVolcano(res,
lab = rownames(res),
x = 'log2(Fold_change)',
y = 'p-value',
xlim = c(-4, 4),
ylim = c(0,15),
title = 'A versus B',
pCutoff = 10e-3,
FCcutoff = 1.5,
transcriptPointSize = 3.0,
transcriptLabSize = 3.0,
#shape = 8, #点形状
shape = c(1, 4, 23, 25), #形状列表
colAlpha = 1)
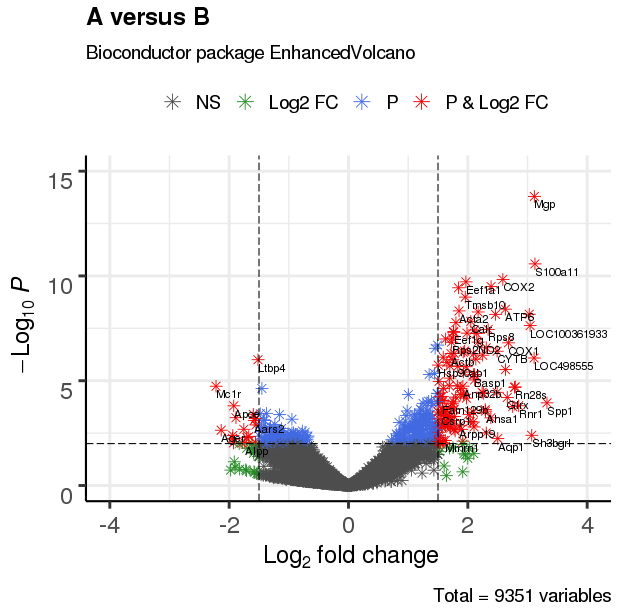
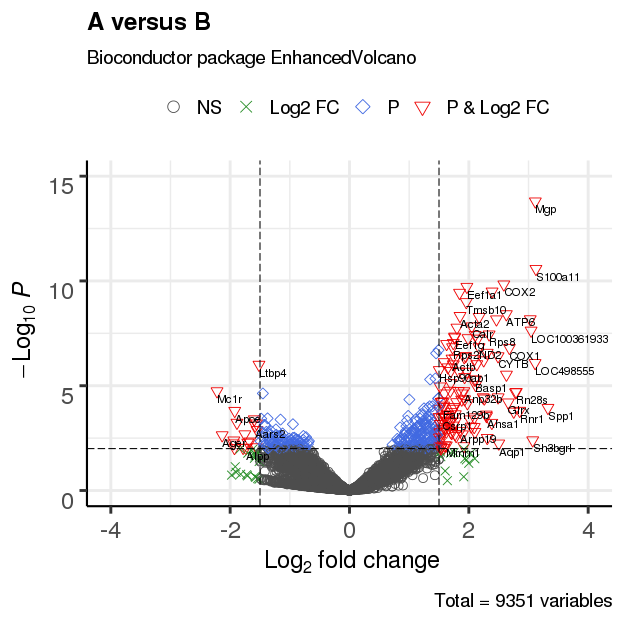
3.4 改变截止线及添加阈值线
EnhancedVolcano(res,
lab = rownames(res),
x = 'log2(Fold_change)',
y = 'p-value',
xlim = c(-4, 4),
ylim = c(0,15),
title = 'A versus B',
pCutoff = 10e-3,
FCcutoff = 1.5,
transcriptPointSize = 3.0,
transcriptLabSize = 3.0,
colAlpha = 1,
cutoffLineType = 'blank',
cutoffLineCol = 'black',
cutoffLineWidth = 0.8,
hline = c(10e-4, 10e-8, 10e-12, 10e-15),
hlineCol = c('grey0', 'grey25','grey50','grey75'),
hlineType = 'longdash',
hlineWidth = 0.8,
gridlines.major = FALSE,
gridlines.minor = FALSE)
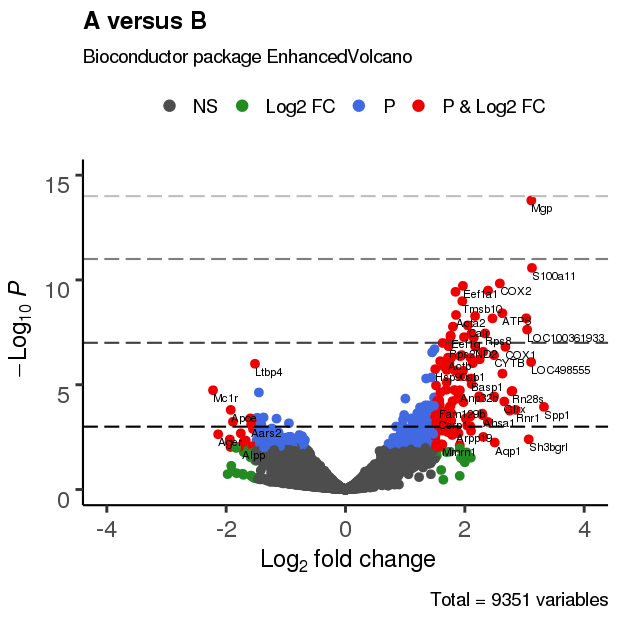
3.5 调整标注位置、大小及文字
EnhancedVolcano(res,
lab = rownames(res),
x = 'log2(Fold_change)',
y = 'p-value',
xlim = c(-4, 4),
ylim = c(0,15),
title = 'A versus B',
pCutoff = 10e-3,
FCcutoff = 1.5,
transcriptPointSize = 3.0,
transcriptLabSize = 3.0,
colAlpha = 1,
cutoffLineType = 'twodash',
cutoffLineWidth = 0.8,
legend=c('NS','Log (base 2) fold-change','P value',
'P value & Log (base 2) fold-change'),
legendPosition = 'right',
legendLabSize = 16,
legendIconSize = 5.0)
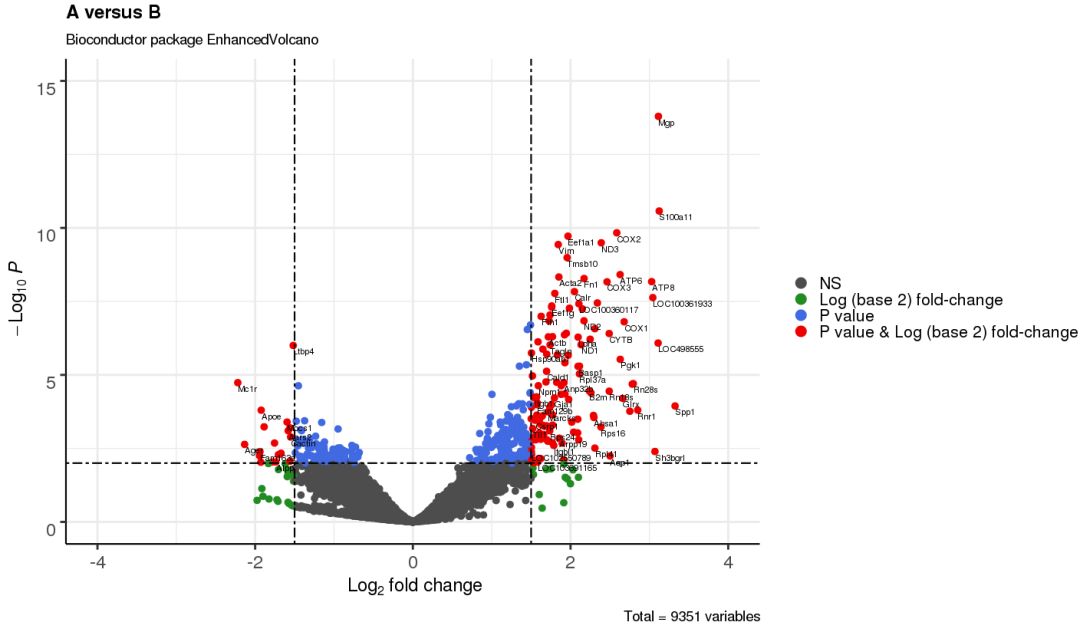
3.6 校正后的 p 值作图
EnhancedVolcano(res,
lab = rownames(res),
x = 'log2(Fold_change)',
y = 'p-value',
xlim = c(-4, 4),
ylim = c(0,15),
title = 'A versus B',
pCutoff = 10e-3,
FCcutoff = 1.5,
xlab = bquote(~Log[2]~ 'fold change'),
ylab = bquote(~-Log[10]~adjusted~italic(P)),
transcriptPointSize = 3.0,
transcriptLabSize = 3.0,
colAlpha = 1,
cutoffLineType = 'twodash',
cutoffLineWidth = 0.8,
legend=c('NS','Log (base 2) fold-change','P value',
'P value & Log (base 2) fold-change'),
legendPosition = 'right',
legendLabSize = 16,
legendIconSize = 5.0)
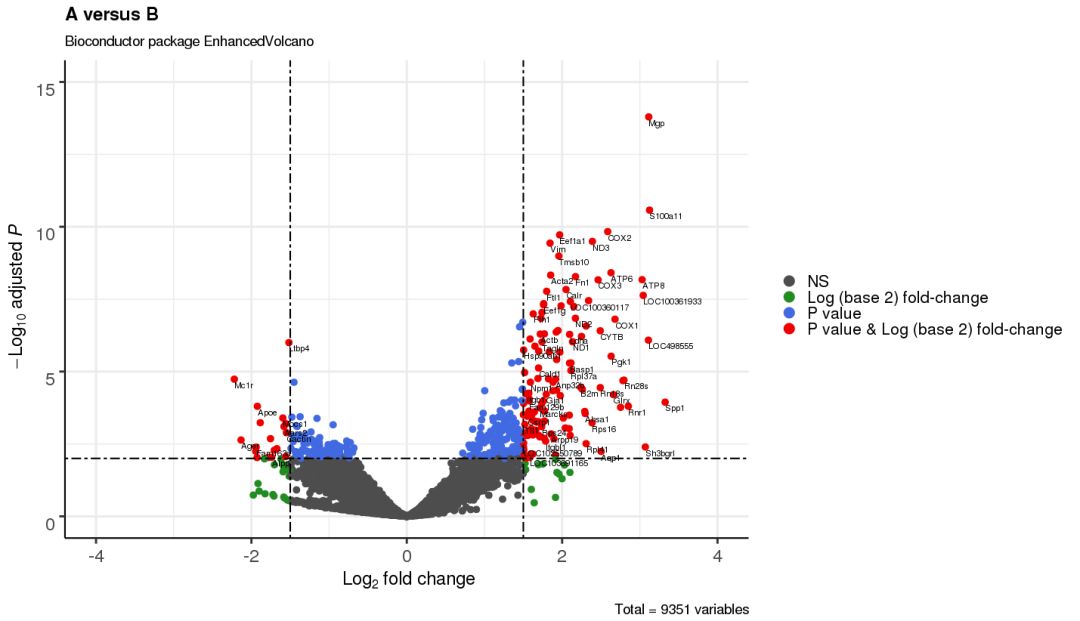
3.7 添加连线用于展示更多标注
EnhancedVolcano(res,
lab = rownames(res),
x = 'log2(Fold_change)',
y = 'p-value',
xlim = c(-4, 4),
ylim = c(0,15),
title = 'A versus B',
pCutoff = 10e-3,
FCcutoff = 1.5,
xlab = bquote(~Log[2]~ 'fold change'),
ylab = bquote(~-Log[10]~adjusted~italic(P)),
transcriptPointSize = 3.0,
transcriptLabSize = 3.0,
colAlpha = 1,
cutoffLineType = 'twodash',
cutoffLineWidth = 0.8,
legend=c('NS','Log (base 2) fold-change','P value',
'P value & Log (base 2) fold-change'),
legendPosition = 'right',
drawConnectors = TRUE,
legendLabSize = 16,
legendIconSize = 5.0)
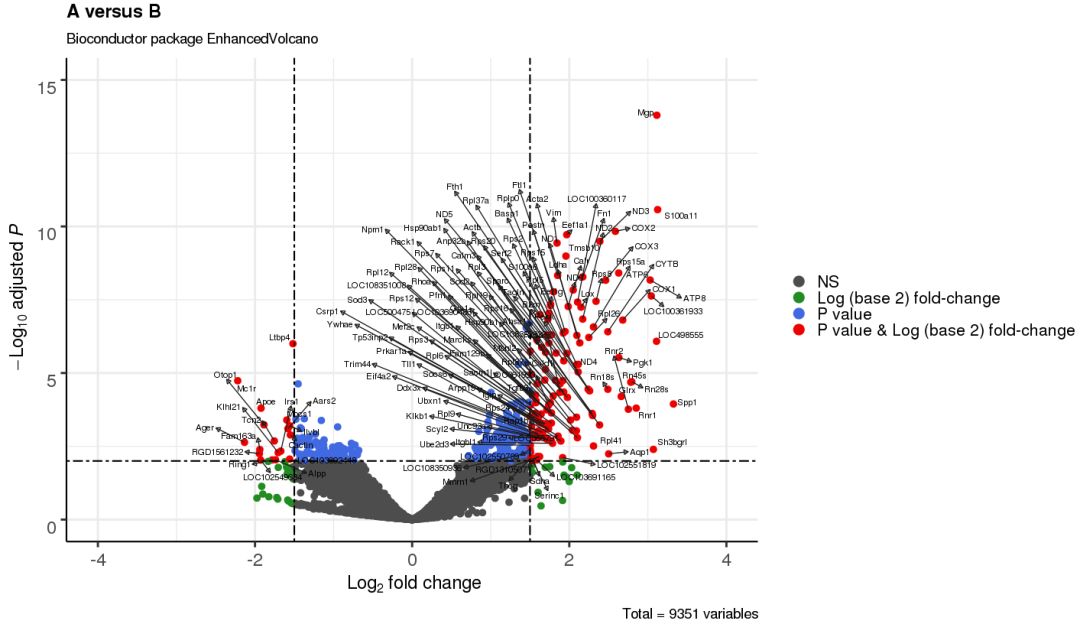
3.8 只标注重要变量
EnhancedVolcano(res,
lab = rownames(res),
x = 'log2(Fold_change)',
y = 'p-value',
xlim = c(-4, 4),
ylim = c(0,15),
title = 'A versus B',
pCutoff = 10e-3,
FCcutoff = 1.5,
xlab = bquote(~Log[2]~ 'fold change'),
ylab = bquote(~-Log[10]~adjusted~italic(P)),
selectLab = c('Spp1','S100a11','Mgp','LOC498555','Sh3bgrl',
'Ring1','Apoe','Tcn2','Ager','Mc1r'),
transcriptPointSize = 3.0,
transcriptLabSize = 3.0,
colAlpha = 1,
cutoffLineType = 'twodash',
cutoffLineWidth = 0.8,
legend=c('NS','Log (base 2) fold-change','P value',
'P value & Log (base 2) fold-change'),
legendPosition = 'right',
drawConnectors = TRUE,
legendLabSize = 16,
legendIconSize = 5.0)
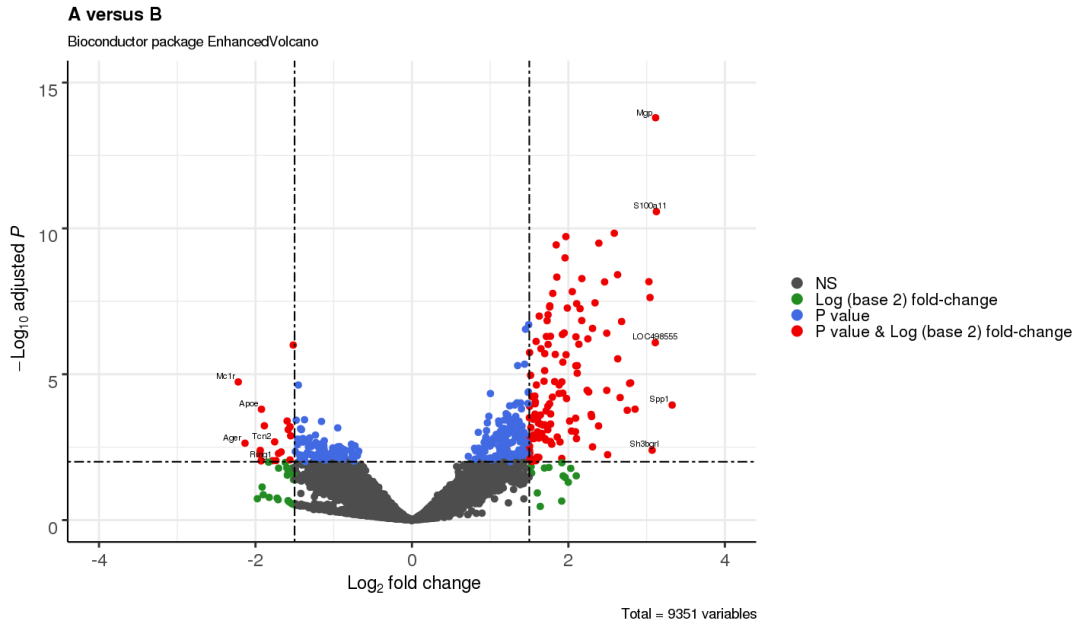
3.9 加框展示变量
EnhancedVolcano(res,
lab = rownames(res),
x = 'log2(Fold_change)',
y = 'p-value',
xlim = c(-4, 4),
ylim = c(0,15),
title = 'A versus B',
pCutoff = 10e-3,
FCcutoff = 1.5,
xlab = bquote(~Log[2]~ 'fold change'),
ylab = bquote(~-Log[10]~adjusted~italic(P)),
selectLab = c('Spp1','S100a11','Mgp','LOC498555','Sh3bgrl',
'Ring1','Apoe','Tcn2','Ager','Mc1r'),
transcriptPointSize = 3.0,
transcriptLabSize = 3.0,
colAlpha = 1,
cutoffLineType = 'twodash',
cutoffLineWidth = 0.8,
legend=c('NS','Log (base 2) fold-change','P value',
'P value & Log (base 2) fold-change'),
legendPosition = 'right',
drawConnectors = TRUE,
boxedlabels = TRUE,
legendLabSize = 16,
legendIconSize = 5.0)
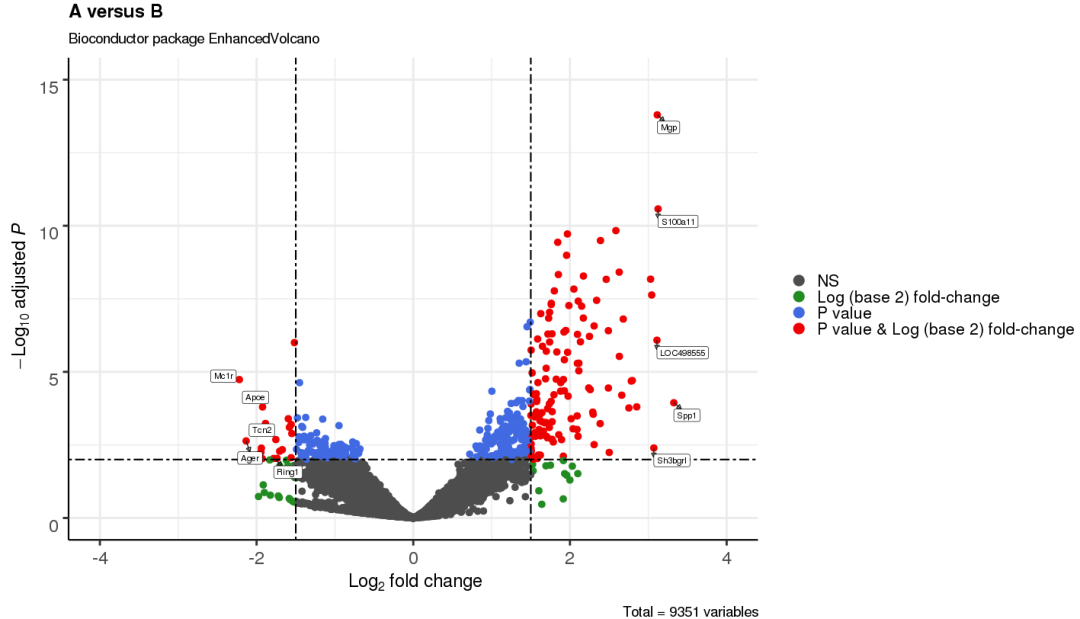
3.10 针对特殊点设置颜色
colCustom 功能可针对特定位点设置颜色,例如上下调基因设置不同颜色,参考代码如下:
keyvals <- rep('black', nrow(res))
# set the base name/label as 'Mid'
names(keyvals) <- rep('Mid', nrow(res))
# fold change > 1.5 & p-value < 0.0001 为高表达
keyvals[which(res$"log2(Fold_change)" > 1.5 & res$"p-value"<0.0001)] <- 'gold'
names(keyvals)[which(res$"log2(Fold_change)" > 1.5 & res$"p-value"<0.0001)] <- 'high'
# fold change < -1.5 & p-value < 0.0001为低表达
keyvals[which(res$"log2(Fold_change)" < -1.5 & res$"p-value"<0.0001)] <- 'royalblue'
names(keyvals)[which(res$"log2(Fold_change)" < -1.5 & res$"p-value"<0.0001)] <- 'low'
EnhancedVolcano(res,
lab = rownames(res),
x = 'log2(Fold_change)',
y = 'p-value',
xlim = c(-4, 4),
ylim = c(0,15),
title = 'A versus B',
pCutoff = 10e-3,
FCcutoff = 1.5,
xlab = bquote(~Log[2]~ 'fold change'),
ylab = bquote(~-Log[10]~adjusted~italic(P)),
selectLab = rownames(res)[which(names(keyvals) %in% c('high', 'low'))],
transcriptPointSize = 3.0,
transcriptLabSize = 3.0,
colAlpha = 1,
cutoffLineType = 'twodash',
cutoffLineWidth = 0.8,
colCustom = keyvals,
border = 'full',
legend=c('NS','Log (base 2) fold-change','P value',
'P value & Log (base 2) fold-change'),
legendPosition = 'right',
drawConnectors = FALSE,
boxedlabels = FALSE,
legendLabSize = 16,
legendIconSize = 5.0)
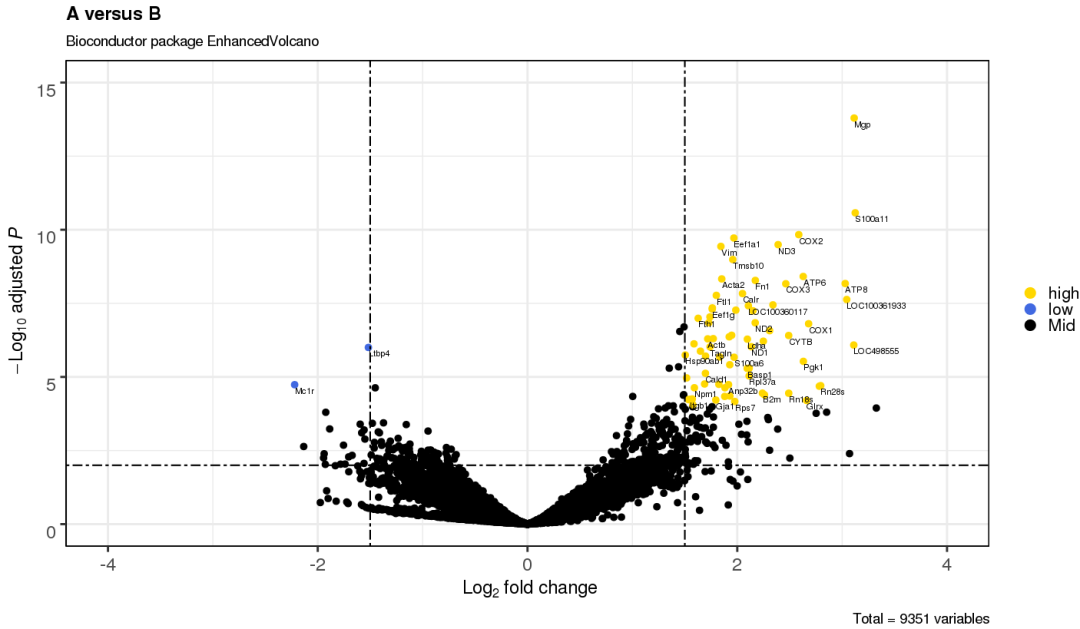
3.11 设置特定点的大小
p <- EnhancedVolcano(res,
lab = rownames(res),
x = 'log2(Fold_change)',
y = 'p-value',
xlim = c(-4, 4),
ylim = c(0,15),
title = '',
subtitle = '',
pCutoff = 10e-3,
FCcutoff = 1.5,
xlab = bquote(~Log[2]~ 'fold change'),
ylab = bquote(~-Log[10]~adjusted~italic(P)),
selectLab = rownames(res)[which(names(keyvals) %in% c('high', 'low'))],
transcriptLabSize = 3.0,
transcriptPointSize = c(ifelse((res$"log2(Fold_change)">2 |res$"log2(Fold_change)"< -2) & res$"p-value"<0.0001 , 3, 1)),
colAlpha = 1,
cutoffLineType = 'twodash',
cutoffLineWidth = 0.8,
colCustom = keyvals,
border = 'full',
legend=c('NS','Log (base 2) fold-change','P value',
'P value & Log (base 2) fold-change'),
legendPosition = 'right',
drawConnectors = FALSE,
boxedlabels = FALSE,
legendLabSize = 16,
legendIconSize = 5.0,
caption = "")
p
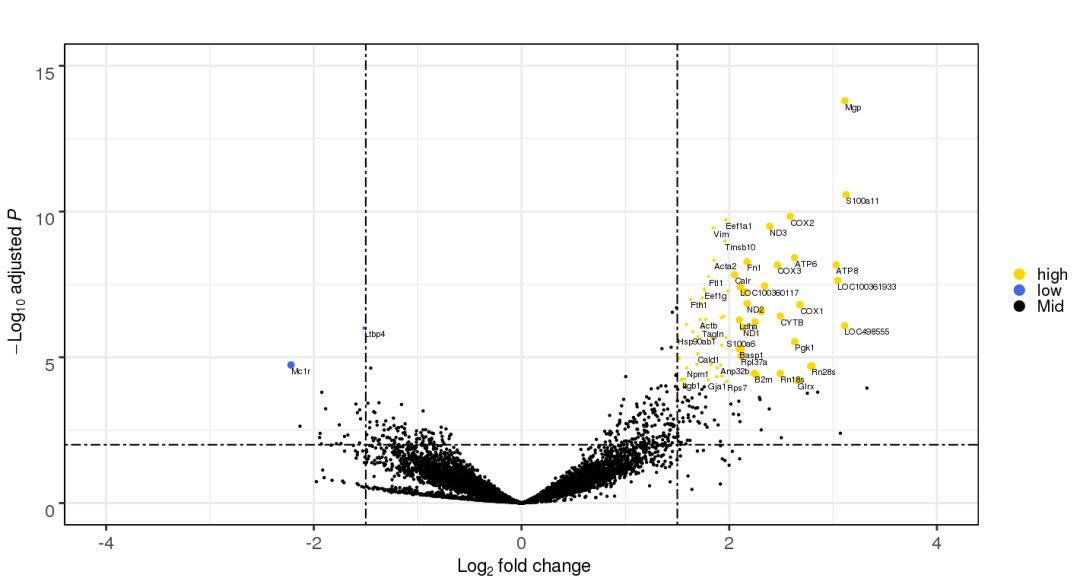
3.12 自定义刻度
p +
ggplot2::coord_cartesian(xlim=c(-6, 6)) +
ggplot2::scale_x_continuous(
breaks=seq(-6,6, 1))
EnhancedVolcano 包绘制火山图就先介绍到这里,如果对你有所帮助,请点个赞吧。

本文分享自微信公众号 - 生信科技爱好者(bioitee)。
如有侵权,请联系 support@oschina.cn 删除。
本文参与“OSC源创计划”,欢迎正在阅读的你也加入,一起分享。


