R 语言绘制环状热图
作者:佳名
来源:简书 - R 语言文集
1. 读取并处理基因表达数据
这是我的基因表达量数据:
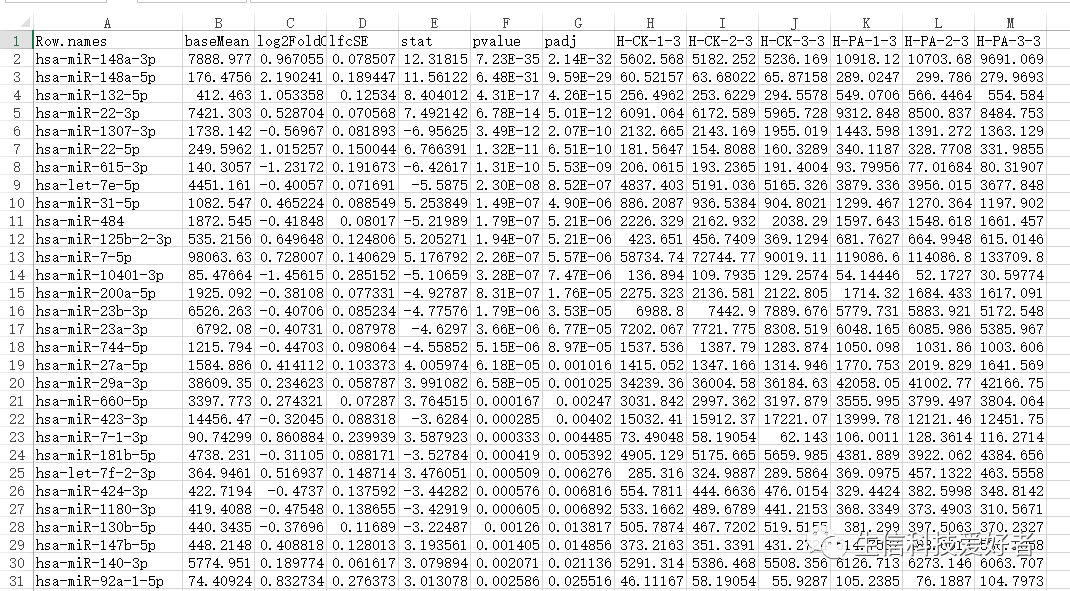
> myfiles <- list.files(pattern = "*.csv")
> myfiles
[1] "4_total_total_FPKM.csv"
> matrix <- read.csv(myfiles[1], sep=',', header=T, check.names=FALSE, row.names=1)
1.1 提取部分数据集
提取 padj 的值小于 0.2 的数据:
> matrix <- subset(matrix, padj<0.2)
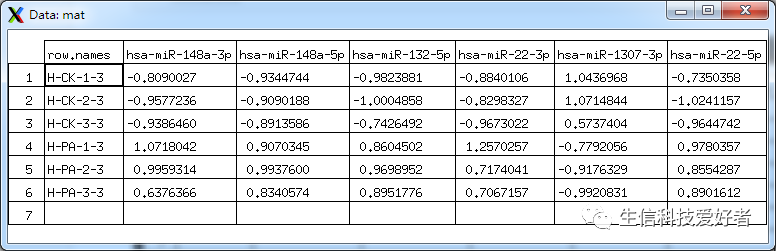
1.2 提取基因表达值所在的列,组成新的矩阵,并将矩阵转置
由于 R 语言的 scale 函数是按列归一化,对于我们一般习惯基因名为行,样本名为列的数据框,就需要进行转置。
mat <- t(matrix[,7:12]) # 7-12 列为每个样本的基因表达量
1.3 基因表达归一化
mat <- scale(mat, center =
TRUE, scale =
TRUE)
View(mat)
1.4 对数据进行聚类,从而得到其 dendrogram
# dist 函数计算 microRNA 间的距离, hclust 函数用来进行层次聚类.
dend <- as.dendrogram(hclust(dist(t(mat))))
1.5 定义进化树颜色
library(dendextend)
n <- 3 # n 可自定义
dend <- dend %>% set("branches_k_color", k = n)
1.6 可视化处理
par(mar=c(7.5,3,1,0))
plot(dend)
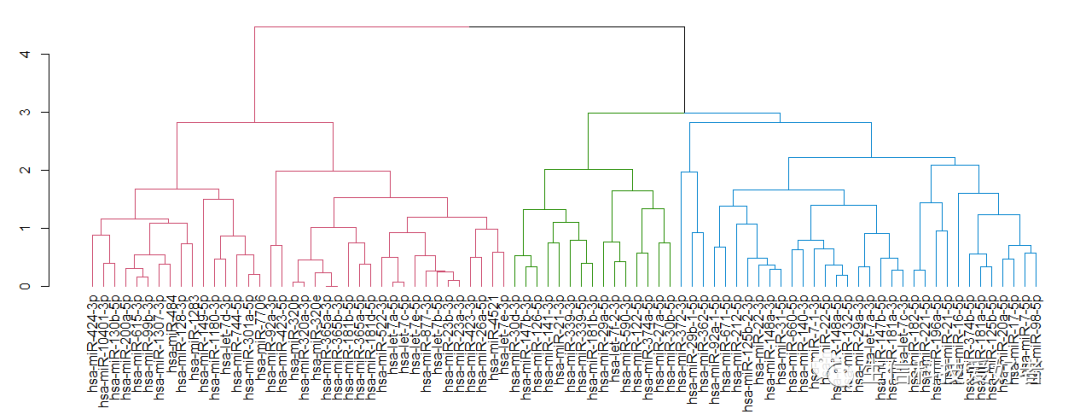
1.7 聚类后的矩阵
如图 Fig 3,聚类后的矩阵的列的顺序会发生变化。按此顺序,重新排列矩阵。
mat2 <- mat[, order.dendrogram(dend)]- 查看矩阵重排后的样本名
> lable1 <- row.names(mat2);
> lable1
[1] "H-CK-1-3" "H-CK-2-3" "H-CK-3-3" "H-PA-1-3" "H-PA-2-3" "H-PA-3-3"
查看矩阵重排后的基因名
> lable2 <- colnames(mat2);
> lable2
[1] "hsa-miR-424-3p" "hsa-miR-10401-3p" "hsa-miR-130b-5p"
[4] "hsa-miR-200a-5p" "hsa-miR-615-3p" "hsa-miR-99b-3p"
[7] "hsa-miR-1307-3p" "hsa-miR-484" "hsa-miR-128-3p"
[10] "hsa-miR-1283" "hsa-miR-149-5p" "hsa-miR-1180-3p"
[13] "hsa-let-7d-3p" "hsa-miR-744-5p" "hsa-miR-301a-5p"
[16] "hsa-miR-7706" "hsa-miR-92a-3p" "hsa-miR-423-5p"
[19] "hsa-miR-320b" "hsa-miR-320a-3p" "hsa-miR-320e"
[22] "hsa-miR-365a-3p" "hsa-miR-365b-3p" "hsa-miR-181b-5p"
[25] "hsa-miR-365a-5p" "hsa-miR-181d-5p" "hsa-miR-522-3p"
[28] "hsa-let-7a-5p" "hsa-let-7c-5p" "hsa-let-7e-5p"
[31] "hsa-miR-877-3p" "hsa-let-7b-5p" "hsa-miR-23b-3p"
[34] "hsa-miR-23a-3p" "hsa-miR-423-3p" "hsa-miR-26a-5p"
[37] "hsa-miR-4521" "hsa-let-7e-3p" "hsa-miR-30d-3p"
[40] "hsa-miR-147b-3p" "hsa-miR-126-5p" "hsa-miR-141-3p"
[43] "hsa-miR-21-3p" "hsa-miR-339-3p" "hsa-miR-339-5p"
[46] "hsa-miR-181b-3p" "hsa-miR-29a-5p" "hsa-let-7f-2-3p"
[49] "hsa-miR-590-3p" "hsa-miR-122-5p" "hsa-miR-374a-5p"
[52] "hsa-miR-27a-5p" "hsa-miR-30b-5p" "hsa-miR-372-3p"
[55] "hsa-miR-29b-1-5p" "hsa-miR-362-5p" "hsa-miR-92a-1-5p"
[58] "hsa-miR-671-5p" "hsa-miR-212-5p" "hsa-miR-125b-2-3p"
[61] "hsa-miR-22-3p" "hsa-miR-148a-3p" "hsa-miR-31-5p"
[64] "hsa-miR-660-5p" "hsa-miR-140-3p" "hsa-miR-7-1-3p"
[67] "hsa-miR-22-5p" "hsa-miR-148a-5p" "hsa-miR-132-5p"
[70] "hsa-miR-29a-3p" "hsa-let-7a-3p" "hsa-miR-147b-5p"
[73] "hsa-miR-181a-3p" "hsa-let-7c-3p" "hsa-miR-182-5p"
[76] "hsa-miR-221-5p" "hsa-miR-196a-5p" "hsa-miR-21-5p"
[79] "hsa-miR-16-5p" "hsa-miR-374b-5p" "hsa-miR-181a-5p"
[82] "hsa-miR-125b-5p" "hsa-miR-20a-5p" "hsa-miR-17-5p"
[85] "hsa-miR-7-5p" "hsa-miR-98-5p"
只有基因名顺序,也就是列名顺序发生了变化。
nr <- nrow(mat2);nr
[1] 6
nc <- ncol(mat2);nc
[1] 86
1.8 构建颜色转变函数
require(
"circlize")
col_fun <- colorRamp2(c(-
1.5,
0,
1.5), c(
"skyblue",
"white",
"red"))
1.9 矩阵中的数值转变为颜色
> col_mat <- col_fun(mat2)
> col_mat[,1] # 查看第一列结果
H-CK-1-3 H-CK-2-3 H-CK-3-3 H-PA-1-3 H-PA-2-3 H-PA-3-3
"#FF0000FF" "#FFDED3FF" "#FFAF96FF" "#ABDBF1FF" "#DCF0F9FF" "#BDE3F4FF"
> col_mat[1,] # 查看第一行的结果
hsa-miR-424-3p hsa-miR-10401-3p hsa-miR-130b-5p hsa-miR-200a-5p
"#FF0000FF" "#FF6645FF" "#FF7B5AFF" "#FF5535FF"
hsa-miR-615-3p hsa-miR-99b-3p hsa-miR-1307-3p hsa-miR-484
"#FF7351FF" "#FF6645FF" "#FF7453FF" "#FF6140FF"
hsa-miR-128-3p hsa-miR-1283 hsa-miR-149-5p hsa-miR-1180-3p
"#FF0000FF" "#FF220EFF" "#FF987AFF" "#FF3B20FF"
hsa-let-7d-3p hsa-miR-744-5p hsa-miR-301a-5p hsa-miR-7706
"#FF2712FF" "#FF1E0CFF" "#FF200DFF" "#FF0000FF"
hsa-miR-92a-3p hsa-miR-423-5p hsa-miR-320b hsa-miR-320a-3p
"#FFA286FF" "#FFAD93FF" "#E4F3FAFF" "#E2F2FAFF"
hsa-miR-320e hsa-miR-365a-3p hsa-miR-365b-3p hsa-miR-181b-5p
"#E1F2FAFF" "#D7EEF8FF" "#D7EEF8FF" "#FFDDD1FF"
hsa-miR-365a-5p hsa-miR-181d-5p hsa-miR-522-3p hsa-let-7a-5p
"#FFECE5FF" "#FBFDFEFF" "#F3FAFDFF" "#FFF2ECFF"
hsa-let-7c-5p hsa-let-7e-5p hsa-miR-877-3p hsa-let-7b-5p
"#FFF6F2FF" "#FFB7A0FF" "#FFC1ACFF" "#FFDED2FF"
hsa-miR-23b-3p hsa-miR-23a-3p hsa-miR-423-3p hsa-miR-26a-5p
"#FFC8B5FF" "#FFD1C1FF" "#FFDACDFF" "#FFDED2FF"
hsa-miR-4521 hsa-let-7e-3p hsa-miR-30d-3p hsa-miR-147b-3p
"#FFA286FF" "#FFAE94FF" "#F0F8FCFF" "#FFE9E1FF"
hsa-miR-126-5p hsa-miR-141-3p hsa-miR-21-3p hsa-miR-339-3p
"#FFDDD1FF" "#E9F5FBFF" "#FAFDFEFF" "#DCF0F9FF"
hsa-miR-339-5p hsa-miR-181b-3p hsa-miR-29a-5p hsa-let-7f-2-3p
"#E3F3FAFF" "#C9E8F6FF" "#95D3EDFF" "#B3DEF2FF"
hsa-miR-590-3p hsa-miR-122-5p hsa-miR-374a-5p hsa-miR-27a-5p
"#B5DFF2FF" "#C5E6F5FF" "#D8EEF8FF" "#D1EBF7FF"
hsa-miR-30b-5p hsa-miR-372-3p hsa-miR-29b-1-5p hsa-miR-362-5p
"#CCE9F6FF" "#D7EDF8FF" "#A1D7EFFF" "#87CEEBFF"
hsa-miR-92a-1-5p hsa-miR-671-5p hsa-miR-212-5p hsa-miR-125b-2-3p
"#AADBF0FF" "#B3DFF2FF" "#C1E4F4FF" "#C0E4F4FF"
hsa-miR-22-3p hsa-miR-148a-3p hsa-miR-31-5p hsa-miR-660-5p
"#BCE2F3FF" "#C2E4F4FF" "#B1DEF2FF" "#B3DFF2FF"
hsa-miR-140-3p hsa-miR-7-1-3p hsa-miR-22-5p hsa-miR-148a-5p
"#A7DAF0FF" "#D4ECF7FF" "#C7E7F5FF" "#B8E0F3FF"
hsa-miR-132-5p hsa-miR-29a-3p hsa-let-7a-3p hsa-miR-147b-5p
"#B4DFF2FF" "#9ED6EFFF" "#9BD5EEFF" "#B2DEF2FF"
hsa-miR-181a-3p hsa-let-7c-3p hsa-miR-182-5p hsa-miR-221-5p
"#AEDCF1FF" "#B3DFF2FF" "#87CEEBFF" "#87CEEBFF"
hsa-miR-196a-5p hsa-miR-21-5p hsa-miR-16-5p hsa-miR-374b-5p
"#A2D8EFFF" "#D2EBF7FF" "#87CEEBFF" "#93D2EDFF"
hsa-miR-181a-5p hsa-miR-125b-5p hsa-miR-20a-5p hsa-miR-17-5p
"#87CEEBFF" "#87CEEBFF" "#87CEEBFF" "#87CEEBFF"
hsa-miR-7-5p hsa-miR-98-5p
"#94D3EDFF" "#87CEEBFF"
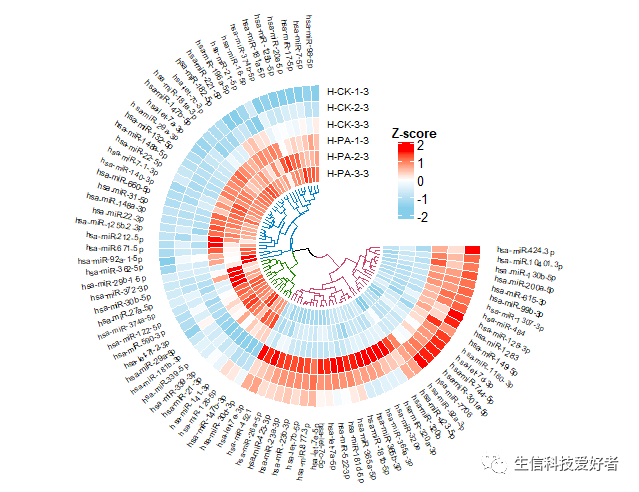
2. 画板设置与绘图
2.1 画板初始化设置
par(mar <- c(0,0,0,0))
circos.clear();
circos.par(canvas.xlim = c(-1.4,1.4),
canvas.ylim = c(-1.4,1.4),
cell.padding = c(0,0,0,0),
gap.degree = 90)
factors <- "a"
circos.initialize(factors, xlim = c(0, ncol(mat2)))
2.2 添加第一个轨道
circos.track(ylim = c(0, nr),bg.border = NA,track.height = 0.1*nr,
panel.fun = function(x, y) {
for(i in 1:nr) {
circos.rect(xleft = 1:nc - 1, ybottom = rep(nr - i, nc),
xright = 1:nc, ytop = rep(nr - i + 1, nc),
border = "white",
col = col_mat[i,])
circos.text(x = nc,
y = 6.4 -i,
labels = lable1[i],
facing = "downward", niceFacing = TRUE,
cex = 0.6,
adj = c(-0.2, 0))
}
})
2.3 添加基因名称
for(i in 1:nc){
circos.text(x = i-0.4,
y = 7,
labels = lable2[i],
facing = "clockwise", niceFacing = TRUE,
cex = 0.5,adj = c(0, 0))
}
2.4 添加进化树
max_height <-max(attr(dend, "height"))
circos.track(ylim = c(0, max_height),bg.border = NA,track.height = 0.3,
panel.fun = function(x, y){
circos.dendrogram(dend = dend,
max_height = max_height)
})
circos.clear()
2.5 添加图例
library(ComplexHeatmap)
lgd <- Legend(at = c(-2,-1, 0, 1, 2), col_fun = col_fun,
title_position = "topcenter",title = "Z-score")
draw(lgd, x = unit(0.65, "npc"), y = unit(0.65, "npc"))
 —
END—
—
END—
声明:本文经原作者同意后授权转载,文章(包括文字和图片)的著作权归原作者所有,任何形式的转载都请联系原作者。

本文分享自微信公众号 - 生信科技爱好者(bioitee)。
如有侵权,请联系 support@oschina.cn 删除。
本文参与“OSC源创计划”,欢迎正在阅读的你也加入,一起分享。


