机器学习sklearn(68):算法实例(二十五)分类(十二)SVM(三)sklearn.svm.SVC(二)
2 非线性SVM与核函数
2.1 SVC在非线性数据上的推广


2.2 重要参数kernel
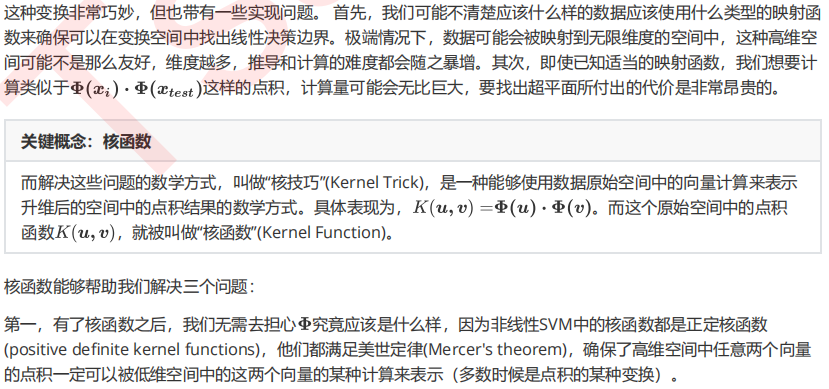



clf = SVC(kernel = "rbf").fit(X,y) plt.scatter(X[:,0],X[:,1],c=y,s=50,cmap="rainbow") plot_svc_decision_function(clf)
可以看到,决策边界被完美地找了出来。
2.3 探索核函数在不同数据集上的表现

1. 导入所需要的库和模块
import numpy as np import matplotlib.pyplot as plt from matplotlib.colors import ListedColormap from sklearn import svm from sklearn.datasets import make_circles, make_moons, make_blobs,make_classification
2. 创建数据集,定义核函数的选择
n_samples = 100 datasets = [ make_moons(n_samples=n_samples, noise=0.2, random_state=0), make_circles(n_samples=n_samples, noise=0.2, factor=0.5, random_state=1), make_blobs(n_samples=n_samples, centers=2, random_state=5), make_classification(n_samples=n_samples,n_features = 2,n_informative=2,n_redundant=0, random_state=5) ] Kernel = ["linear","poly","rbf","sigmoid"]
#四个数据集分别是什么样子呢? for X,Y in datasets: plt.figure(figsize=(5,4)) plt.scatter(X[:,0],X[:,1],c=Y,s=50,cmap="rainbow")

3. 构建子图
nrows=len(datasets) ncols=len(Kernel) + 1 fig, axes = plt.subplots(nrows, ncols,figsize=(20,16))
4. 开始进行子图循环
#第一层循环:在不同的数据集中循环 for ds_cnt, (X,Y) in enumerate(datasets): #在图像中的第一列,放置原数据的分布 ax = axes[ds_cnt, 0] if ds_cnt == 0: ax.set_title("Input data") ax.scatter(X[:, 0], X[:, 1], c=Y, zorder=10, cmap=plt.cm.Paired,edgecolors='k') ax.set_xticks(()) ax.set_yticks(()) #第二层循环:在不同的核函数中循环 #从图像的第二列开始,一个个填充分类结果 for est_idx, kernel in enumerate(Kernel): #定义子图位置 ax = axes[ds_cnt, est_idx + 1] #建模 clf = svm.SVC(kernel=kernel, gamma=2).fit(X, Y) score = clf.score(X, Y) #绘制图像本身分布的散点图 ax.scatter(X[:, 0], X[:, 1], c=Y ,zorder=10 ,cmap=plt.cm.Paired,edgecolors='k') #绘制支持向量 ax.scatter(clf.support_vectors_[:, 0], clf.support_vectors_[:, 1], s=50, facecolors='none', zorder=10, edgecolors='k') #绘制决策边界 x_min, x_max = X[:, 0].min() - .5, X[:, 0].max() + .5 y_min, y_max = X[:, 1].min() - .5, X[:, 1].max() + .5 #np.mgrid,合并了我们之前使用的np.linspace和np.meshgrid的用法 #一次性使用最大值和最小值来生成网格 #表示为[起始值:结束值:步长] #如果步长是复数,则其整数部分就是起始值和结束值之间创建的点的数量,并且结束值被包含在内 XX, YY = np.mgrid[x_min:x_max:200j, y_min:y_max:200j] #np.c_,类似于np.vstack的功能 Z = clf.decision_function(np.c_[XX.ravel(), YY.ravel()]).reshape(XX.shape) #填充等高线不同区域的颜色 ax.pcolormesh(XX, YY, Z > 0, cmap=plt.cm.Paired) #绘制等高线 ax.contour(XX, YY, Z, colors=['k', 'k', 'k'], linestyles=['--', '-', '--'], levels=[-1, 0, 1]) #设定坐标轴为不显示 ax.set_xticks(()) ax.set_yticks(()) #将标题放在第一行的顶上 if ds_cnt == 0: ax.set_title(kernel) #为每张图添加分类的分数 ax.text(0.95, 0.06, ('%.2f' % score).lstrip('0') , size=15 , bbox=dict(boxstyle='round', alpha=0.8, facecolor='white') #为分数添加一个白色的格子作为底色 , transform=ax.transAxes #确定文字所对应的坐标轴,就是ax子图的坐标轴本身 , horizontalalignment='right' #位于坐标轴的什么方向 ) plt.tight_layout() plt.show()


2.4 探索核函数的优势和缺陷
看起来,除了Sigmoid核函数,其他核函数效果都还不错。但其实rbf和poly都有自己的弊端,我们使用乳腺癌数据集作为例子来展示一下:
from sklearn.datasets import load_breast_cancer from sklearn.svm import SVC from sklearn.model_selection import train_test_split import matplotlib.pyplot as plt import numpy as np from time import time import datetime data = load_breast_cancer() X = data.data y = data.target X.shape np.unique(y) plt.scatter(X[:,0],X[:,1],c=y) plt.show() Xtrain, Xtest, Ytrain, Ytest = train_test_split(X,y,test_size=0.3,random_state=420) Kernel = ["linear","poly","rbf","sigmoid"] for kernel in Kernel: time0 = time() clf= SVC(kernel = kernel , gamma="auto" # , degree = 1 , cache_size=5000 ).fit(Xtrain,Ytrain) print("The accuracy under kernel %s is %f" % (kernel,clf.score(Xtest,Ytest))) print(datetime.datetime.fromtimestamp(time()-time0).strftime("%M:%S:%f"))

Kernel = ["linear","rbf","sigmoid"] for kernel in Kernel: time0 = time() clf= SVC(kernel = kernel , gamma="auto" # , degree = 1 , cache_size=5000 ).fit(Xtrain,Ytrain) print("The accuracy under kernel %s is %f" % (kernel,clf.score(Xtest,Ytest))) print(datetime.datetime.fromtimestamp(time()-time0).strftime("%M:%S:%f"))

Kernel = ["linear","poly","rbf","sigmoid"] for kernel in Kernel: time0 = time() clf= SVC(kernel = kernel , gamma="auto" , degree = 1 , cache_size=5000 ).fit(Xtrain,Ytrain) print("The accuracy under kernel %s is %f" % (kernel,clf.score(Xtest,Ytest))) print(datetime.datetime.fromtimestamp(time()-time0).strftime("%M:%S:%f"))

import pandas as pd data = pd.DataFrame(X) data.describe([0.01,0.05,0.1,0.25,0.5,0.75,0.9,0.99]).T
望去,果然数据存在严重的量纲不一的问题。我们来使用数据预处理中的标准化的类,对数据进行标准化:
from sklearn.preprocessing import StandardScaler X = StandardScaler().fit_transform(X) data = pd.DataFrame(X) data.describe([0.01,0.05,0.1,0.25,0.5,0.75,0.9,0.99]).T
标准化完毕后,再次让SVC在核函数中遍历,此时我们把degree的数值设定为1,观察各个核函数在去量纲后的数据上的表现:
Xtrain, Xtest, Ytrain, Ytest = train_test_split(X,y,test_size=0.3,random_state=420) Kernel = ["linear","poly","rbf","sigmoid"] for kernel in Kernel: time0 = time() clf= SVC(kernel = kernel , gamma="auto" , degree = 1 , cache_size=5000 ).fit(Xtrain,Ytrain) print("The accuracy under kernel %s is %f" % (kernel,clf.score(Xtest,Ytest))) print(datetime.datetime.fromtimestamp(time()-time0).strftime("%M:%S:%f"))

2.5 选取与核函数相关的参数:degree & gamma & coef0



score = [] gamma_range = np.logspace(-10, 1, 50) #返回在对数刻度上均匀间隔的数字 for i in gamma_range: clf = SVC(kernel="rbf",gamma = i,cache_size=5000).fit(Xtrain,Ytrain) score.append(clf.score(Xtest,Ytest)) print(max(score), gamma_range[score.index(max(score))]) plt.plot(gamma_range,score) plt.show()

from sklearn.model_selection import StratifiedShuffleSplit from sklearn.model_selection import GridSearchCV time0 = time() gamma_range = np.logspace(-10,1,20) coef0_range = np.linspace(0,5,10) param_grid = dict(gamma = gamma_range ,coef0 = coef0_range) cv = StratifiedShuffleSplit(n_splits=5, test_size=0.3, random_state=420) grid = GridSearchCV(SVC(kernel = "poly",degree=1,cache_size=5000), param_grid=param_grid, cv=cv) grid.fit(X, y) print("The best parameters are %s with a score of %0.5f" % (grid.best_params_, grid.best_score_)) print(datetime.datetime.fromtimestamp(time()-time0).strftime("%M:%S:%f"))

3 硬间隔与软间隔:重要参数C
3.1 SVM在软间隔数据上的推广
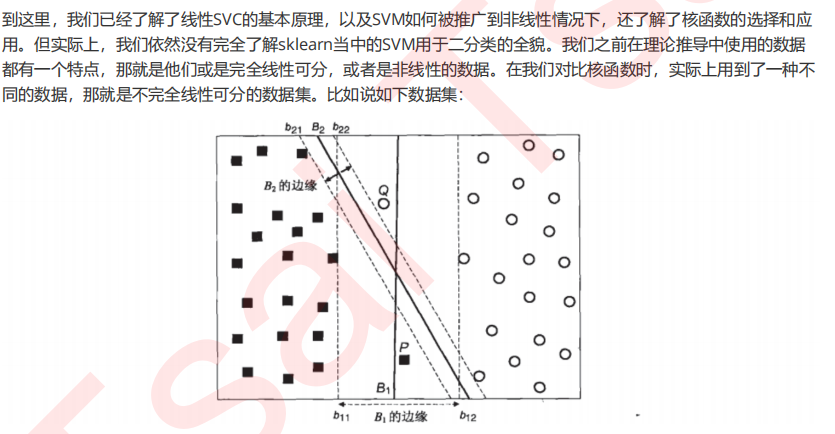


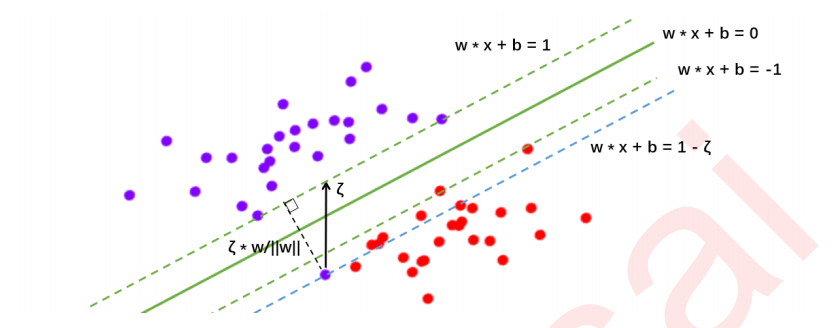


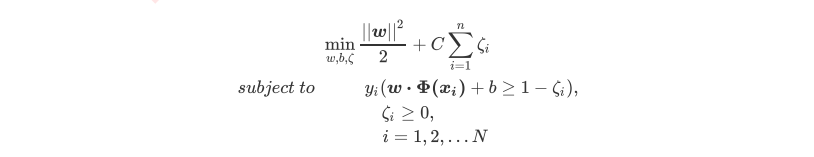

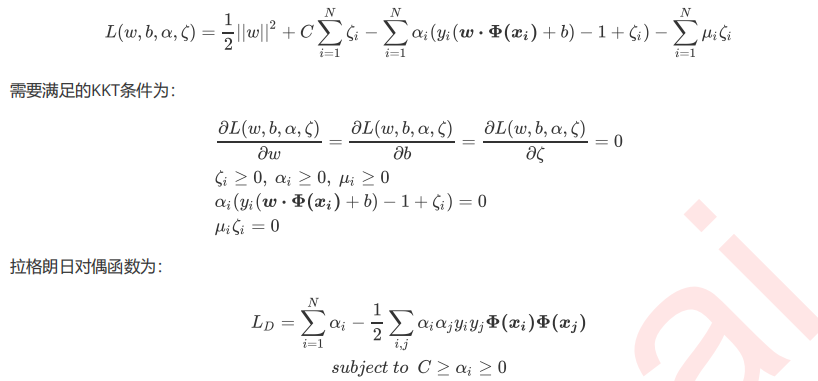

3.2 重要参数C
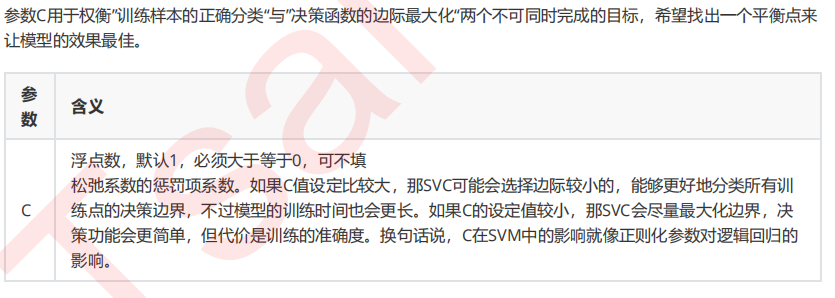

#调线性核函数 score = [] C_range = np.linspace(0.01,30,50) for i in C_range: clf = SVC(kernel="linear",C=i,cache_size=5000).fit(Xtrain,Ytrain) score.append(clf.score(Xtest,Ytest)) print(max(score), C_range[score.index(max(score))]) plt.plot(C_range,score) plt.show() #换rbf score = [] C_range = np.linspace(0.01,30,50) for i in C_range: clf = SVC(kernel="rbf",C=i,gamma = 0.012742749857031322,cache_size=5000).fit(Xtrain,Ytrain) score.append(clf.score(Xtest,Ytest)) print(max(score), C_range[score.index(max(score))]) plt.plot(C_range,score) plt.show() #进一步细化 score = [] C_range = np.linspace(5,7,50) for i in C_range: clf = SVC(kernel="rbf",C=i,gamma = 0.012742749857031322,cache_size=5000).fit(Xtrain,Ytrain) score.append(clf.score(Xtest,Ytest)) print(max(score), C_range[score.index(max(score))]) plt.plot(C_range,score) plt.show()

4 总结

本文来自博客园,作者:秋华,转载请注明原文链接:https://www.cnblogs.com/qiu-hua/p/14952554.html


