引言
在进行科学计算的过程的中,很多时候我们需要将中间的计算结果保存下来以便后续查看,或者还需要后续再进行分析处理。
解决方案
1、一维/二维数组保存/载入
特别地,针对一维/二维数组保存,numpy自带savetxt函数有实现;针对保存在txt文件中的数组,numpy自带loadtxt函数。接下来先简单记录savetxt/loadtxt函数参数说明,再举例说明。
numpy.savetxt(fname, X, fmt='%.18e', delimiter=' ', newline='n', header='', footer='', comments='# ', encoding=None)
Save an array to a text file.
| Parameters: |
- fname : filename or file handle
-
If the filename ends in .gz, the file is automatically saved in compressed gzip format. loadtxtunderstands gzipped files transparently.
- X : 1D or 2D array_like
-
Data to be saved to a text file.
- fmt : str or sequence of strs, optional
-
A single format (%10.5f), a sequence of formats, or a multi-format string, e.g. ‘Iteration %d – %10.5f’, in which case delimiter is ignored. For complex X, the legal options for fmt are:
- a single specifier, fmt=’%.4e’, resulting in numbers formatted like ‘ (%s+%sj)’ % (fmt, fmt)
- a full string specifying every real and imaginary part, e.g. ‘ %.4e %+.4ej %.4e %+.4ej %.4e %+.4ej’ for 3 columns
- a list of specifiers, one per column - in this case, the real and imaginary part must have separate specifiers, e.g. [‘%.3e + %.3ej’, ‘(%.15e%+.15ej)’] for 2 columns
- delimiter : str, optional
-
String or character separating columns.
- newline : str, optional
-
String or character separating lines.
- header : str, optional
-
String that will be written at the beginning of the file.
- footer : str, optional
-
String that will be written at the end of the file.
- comments : str, optional
-
String that will be prepended to the header and footer strings, to mark them as comments. Default: ‘# ‘, as expected by e.g. numpy.loadtxt.
- encoding : {None, str}, optional
-
Encoding used to encode the outputfile. Does not apply to output streams. If the encoding is something other than ‘bytes’ or ‘latin1’ you will not be able to load the file in NumPy versions < 1.14. Default is ‘latin1’.
|
|---|
说明:比较常用的参数:fname: 保存的文件名,X: 需要保存的数据,fmt:格式化, delimiter:列之间的分隔串,newline:行之间的分隔串
Further explanation of the fmt parameter (%[flag]width[.precision]specifier):
- flags:
-
- : left justify
+ : Forces to precede result with + or -.
0 : Left pad the number with zeros instead of space (see width).
- width:
- Minimum number of characters to be printed. The value is not truncated if it has more characters.
- precision:
-
- For integer specifiers (eg.
d,i,o,x), the minimum number of digits.
- For
e, E and f specifiers, the number of digits to print after the decimal point.
- For
g and G, the maximum number of significant digits.
- For
s, the maximum number of characters.
- specifiers:
-
c : character
d or i : signed decimal integer
e or E : scientific notation with e or E.
f : decimal floating point
g,G : use the shorter of e,E or f
o : signed octal
s : string of characters
u : unsigned decimal integer
x,X : unsigned hexadecimal integer
This explanation of fmt is not complete, for an exhaustive specification see [1].
例子:
1 import numpy as np
2
3 a = np.random.randn(3, 5).astype(np.float32)
4
5 np.savetxt('./a.txt', a, fmt='%2.5f', delimiter='s')
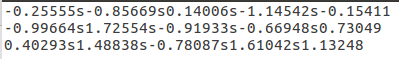
‘a.txt'文件中的内容:
numpy.loadtxt(fname, dtype=<class 'float'>, comments='#', delimiter=None, converters=None, skiprows=0, usecols=None, unpack=False, ndmin=0, encoding='bytes', max_rows=None)
Load data from a text file.
Each row in the text file must have the same number of values.
| Parameters: |
- fname : file, str, or pathlib.Path
-
File, filename, or generator to read. If the filename extension is .gz or .bz2, the file is first decompressed. Note that generators should return byte strings for Python 3k.
- dtype : data-type, optional
-
Data-type of the resulting array; default: float. If this is a structured data-type, the resulting array will be 1-dimensional, and each row will be interpreted as an element of the array. In this case, the number of columns used must match the number of fields in the data-type.
- comments : str or sequence of str, optional
-
The characters or list of characters used to indicate the start of a comment. None implies no comments. For backwards compatibility, byte strings will be decoded as ‘latin1’. The default is ‘#’.
- delimiter : str, optional
-
The string used to separate values. For backwards compatibility, byte strings will be decoded as ‘latin1’. The default is whitespace.
- converters : dict, optional
-
A dictionary mapping column number to a function that will parse the column string into the desired value. E.g., if column 0 is a date string: converters = {0: datestr2num}. Converters can also be used to provide a default value for missing data (but see also genfromtxt): converters = {3: lambda s: float(s.strip() or 0)}. Default: None.
- skiprows : int, optional
-
Skip the first skiprows lines, including comments; default: 0.
- usecols : int or sequence, optional
-
Which columns to read, with 0 being the first. For example, usecols = (1,4,5) will extract the 2nd, 5th and 6th columns. The default, None, results in all columns being read.
Changed in version 1.11.0: When a single column has to be read it is possible to use an integer instead of a tuple. E.g usecols = 3 reads the fourth column the same way as usecols = (3,) would.
- unpack : bool, optional
-
If True, the returned array is transposed, so that arguments may be unpacked using x, y, z = loadtxt(...). When used with a structured data-type, arrays are returned for each field. Default is False.
- ndmin : int, optional
-
The returned array will have at least ndmin dimensions. Otherwise mono-dimensional axes will be squeezed. Legal values: 0 (default), 1 or 2.
- encoding : str, optional
-
Encoding used to decode the inputfile. Does not apply to input streams. The special value ‘bytes’ enables backward compatibility workarounds that ensures you receive byte arrays as results if possible and passes ‘latin1’ encoded strings to converters. Override this value to receive unicode arrays and pass strings as input to converters. If set to None the system default is used. The default value is ‘bytes’.
- max_rows : int, optional
-
Read max_rows lines of content after skiprows lines. The default is to read all the lines.
|
|---|
| Returns: |
- out : ndarray
-
Data read from the text file.
|
|---|
例子:
>>> from io import StringIO # StringIO behaves like a file object
>>> c = StringIO(u"0 1\n2 3")
>>> np.loadtxt(c)
array([[0., 1.],
[2., 3.]])
>>> d = StringIO(u"M 21 72\nF 35 58")
>>> np.loadtxt(d, dtype={'names': ('gender', 'age', 'weight'),
... 'formats': ('S1', 'i4', 'f4')})
array([(b'M', 21, 72.), (b'F', 35, 58.)],
dtype=[('gender', 'S1'), ('age', '<i4'), ('weight', '<f4')])
>>> c = StringIO(u"1,0,2\n3,0,4")
>>> x, y = np.loadtxt(c, delimiter=',', usecols=(0, 2), unpack=True)
>>> x
array([1., 3.])
>>> y
array([2., 4.])
2.多维数组的保存和载入
一维/二维数组也可以通过此方式保存/载入,因为方便直接看,所以放在txt文件中。
numpy.save(file, arr, allow_pickle=True, fix_imports=True)[source]
Save an array to a binary file in NumPy .npy format.
| Parameters: |
- file : file, str, or pathlib.Path
-
File or filename to which the data is saved. If file is a file-object, then the filename is unchanged. If file is a string or Path, a .npy extension will be appended to the file name if it does not already have one.
- arr : array_like
-
Array data to be saved.
- allow_pickle : bool, optional
-
Allow saving object arrays using Python pickles. Reasons for disallowing pickles include security (loading pickled data can execute arbitrary code) and portability (pickled objects may not be loadable on different Python installations, for example if the stored objects require libraries that are not available, and not all pickled data is compatible between Python 2 and Python 3). Default: True
- fix_imports : bool, optional
-
Only useful in forcing objects in object arrays on Python 3 to be pickled in a Python 2 compatible way. If fix_imports is True, pickle will try to map the new Python 3 names to the old module names used in Python 2, so that the pickle data stream is readable with Python 2.
-
|
|---|
例子:
1 import numpy as np
2
3 a = np.random.randn(3, 5).astype(np.float32)
4
5 np.save('txt.npy', a)
numpy.load(file, mmap_mode=None, allow_pickle=False, fix_imports=True, encoding='ASCII')[source]
Load arrays or pickled objects from .npy, .npz or pickled files.
Warning
Loading files that contain object arrays uses the pickle module, which is not secure against erroneous or maliciously constructed data. Consider passing allow_pickle=False to load data that is known not to contain object arrays for the safer handling of untrusted sources.
| Parameters: |
- file : file-like object, string, or pathlib.Path
-
The file to read. File-like objects must support the seek() and read() methods. Pickled files require that the file-like object support the readline() method as well.
- mmap_mode : {None, ‘r+’, ‘r’, ‘w+’, ‘c’}, optional
-
If not None, then memory-map the file, using the given mode (see numpy.memmap for a detailed description of the modes). A memory-mapped array is kept on disk. However, it can be accessed and sliced like any ndarray. Memory mapping is especially useful for accessing small fragments of large files without reading the entire file into memory.
- allow_pickle : bool, optional
-
Allow loading pickled object arrays stored in npy files. Reasons for disallowing pickles include security, as loading pickled data can execute arbitrary code. If pickles are disallowed, loading object arrays will fail. Default: False
Changed in version 1.16.3: Made default False in response to CVE-2019-6446.
- fix_imports : bool, optional
-
Only useful when loading Python 2 generated pickled files on Python 3, which includes npy/npz files containing object arrays. If fix_imports is True, pickle will try to map the old Python 2 names to the new names used in Python 3.
- encoding : str, optional
-
What encoding to use when reading Python 2 strings. Only useful when loading Python 2 generated pickled files in Python 3, which includes npy/npz files containing object arrays. Values other than ‘latin1’, ‘ASCII’, and ‘bytes’ are not allowed, as they can corrupt numerical data. Default: ‘ASCII’
|
|---|
| Returns: |
- result : array, tuple, dict, etc.
-
Data stored in the file. For .npz files, the returned instance of NpzFile class must be closed to avoid leaking file descriptors.
|
|---|
| Raises: |
- IOError
-
If the input file does not exist or cannot be read.
- ValueError
-
The file contains an object array, but allow_pickle=False given.
|
|---|
Notes
-
If the file contains pickle data, then whatever object is stored in the pickle is returned.
-
If the file is a .npy file, then a single array is returned.
-
If the file is a .npz file, then a dictionary-like object is returned, containing {filename: array} key-value pairs, one for each file in the archive.
-
If the file is a .npz file, the returned value supports the context manager protocol in a similar fashion to the open function:
with load('foo.npz') as data:
a = data['a']
The underlying file descriptor is closed when exiting the ‘with’ block.
例子:
Store data to disk, and load it again:
>>> np.save('/tmp/123', np.array([[1, 2, 3], [4, 5, 6]]))
>>> np.load('/tmp/123.npy')
array([[1, 2, 3],
[4, 5, 6]])
Store compressed data to disk, and load it again:
>>> a=np.array([[1, 2, 3], [4, 5, 6]])
>>> b=np.array([1, 2])
>>> np.savez('/tmp/123.npz', a=a, b=b)
>>> data = np.load('/tmp/123.npz')
>>> data['a']
array([[1, 2, 3],
[4, 5, 6]])
>>> data['b']
array([1, 2])
>>> data.close()
Mem-map the stored array, and then access the second row directly from disk:
>>> X = np.load('/tmp/123.npy', mmap_mode='r')
>>> X[1, :]
memmap([4, 5, 6])



