统计计算——Bootstrap总结整理
Bootstrapping
Boostrap 有放回的重抽样。
符号定义: 重复抽样的bootstrap
真实分布
Eg.
有一个要做估计的参数
用原始样本做估计的估计值
用重抽样样本做估计的估计值
重复抽样的bootstrap
Bootstrapping generates an empirical distribution of a test statistic or
set of test statistics. 有放回的重抽样。 Advantages:It allows you to
generate confidence intervals and test statistical hypotheses without
having to assume a specific underlying theoretical distribution.
- Randomly select 10 observations from the sample, with replacement
after each selection. Some observations may be selected more than
once, and some may not be selected at all. - Calculate and record the sample mean.
- Repeat steps 1 and 2 a thousand times.
- Order the 1,000 sample means from smallest to largest.
- Find the sample means representing the 2.5th and 97.5th percentiles.
In this case, it's the 25th number from the bottom and top. These
are your 95 percent confidence limits
What if you wanted confidence intervals for the sample median, or
the difference between two sample medians?
When to use ? - the underlying distributions are unknown - outliers
problems - sample sizes are small - parametric approach don't exist.
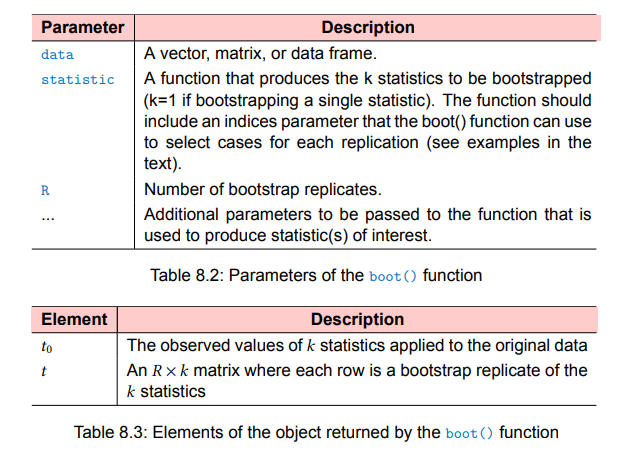
Bootstrapping with R
- Write a function that returns the statistic or statistics of
interest. If there is a single statistic (for example, a median),
the function should return a number. If there is a set of statistics
(for example, a set of regression coefficients), the function should
return a vector. - Process this function through the
boot()function in order to
generate R bootstrap replications of the statistic(s). - Use the
boot.ci()function to obtain confidence intervals for the
statistic(s) generated in step 2.
library(boot)
#bootobject <- boot(data=, statistic =, R=, ...)
#boot.ci(bootobject , conf= ..., type = ...)
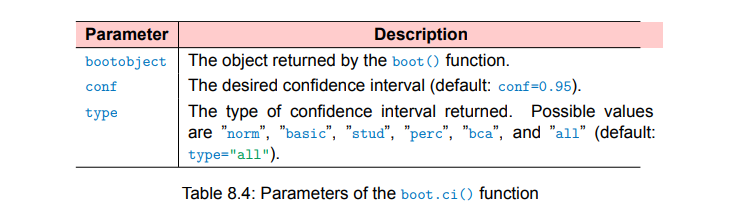
The
bcaprovides an interval that makes simple adjustments for bias.
I findbcapreferable in most circumstances.
#indices <- c[3,1,4,1]
# data[c(3,1,4,1)]
my_mean <- function(x,indices){#这个函数的返回值:要估计的统计量。
mean(x[indices])
}
x <- iris$Petal.Length
library(boot)
boot(x, statistic = my_mean, R = 1000)
- std. error: std of
- bias: 重抽样样本均值估计值和原始抽样值的差异。
- original:
Bootstrapping for an arbitrary parameter
Eg. Financial assets
Bootstrap for an arbitrary parameter
Example (Financial assets)
- Suppose that we wish to invest a fixed sum of money in two financial assets that yield returns of
- Invest a fraction
- Minimize
and we have
where
Question:
How to estimate
Solution:
Solution
- In reality,
- Compute estimates for these quantities:
- Estimate
Estimate the accuracy of
If we know the distribution of
- Resample
- Then, the estimtor for
- The estimator for the standard deviation of
If we do not know the distribution of
- Usually, we don't know the distribution of
- Then, the estimtor for
- The estimator for the standard deviation of
总体的
问题在于,这个估计是有偏的,因为除了一些东西在下面,就不是线性的了。
注意在抽样时X,Y两者是一一对应的。
偏差的计算:
两页ppt上的公式s里面都少了个平方。这里补上了。
alpha.fn = function(data, index) {
X = data$X[index]
Y = data$Y[index]
alpha.hat = (var(Y) - cov(X, Y)) / (var(X) + var(Y) - 2 * cov(X, Y))
return(alpha.hat)
}
library(ISLR2)
alpha.fn(Portfolio, 1:100) # alpha.hat
set.seed(1)
alpha.fn(Portfolio, sample(100, 100, replace = TRUE)) # alpha.hat* # run R=1000 times, we will have E(alpha.hat)
library(boot)
boot.model = boot(Portfolio, alpha.fn, R = 1000)
boot.model
Estimating the accuracy of a linear regression model
summary(lm(mpg ~ horsepower, data = Auto))$coef
coef.fn = function(data, index) {
coef(lm(mpg ~ horsepower, data = data, subset = index))
}
想知道std是否正确,因为
coef.fn = function(data, index) {
coef(lm(mpg ~ horsepower, data = data, subset = index))
}
coef.fn(Auto, 1:392)
set.seed(2)
coef.fn(Auto, sample(392, 392, replace = TRUE))
boot.lm = boot(Auto, coef.fn, 1000)
boot.lm
Question: lm的结果和boot的结果应该相信哪一个?
相信boot的结果,因为lm是参数方法,需要满足一些假设,但是boot是非参方法,不需要数据符合什么假设。
但是如果所有正态(参数)的假设都满足,肯定还是参数的好,但是只要(参数)检验的假设不能满足,那肯定是非参的更好。
t-based bootstrap confidence interval
t-based bootstrap confidence interval
- Let
- Let
- Under most circumstances, as the sample size
Consistency:
Normality??:
- Under this assumption, an approximate
If
很多时候
Complete Bootstrap Estimation
在上述参数估计难以给出时,可以完全用非参的方法。从所有重采样的样本中去找一个下分位点处的样本,和一个上分位点处的样本。
The Bootstrap Estimate of Bias
The Bootstrap Estimate of Bias
- Bootstrapping can also be used to estimate the bias of
- Let
- The Bias of
- Let
- Let
- Based on the Bootstrap method, we can generated
where
- The bias-corrected estimate of
Eg.
unbiased:
biased:
n = 20
set.seed(2022)
data = rnorm(n, 0, 1)
variance = sum((data - mean(data))^2)/n
variance
boots = rep(0,1000)
for(b in 1:1000){
data_b <- sample(data, n, replace=T)
boots[b] = sum((data_b - mean(data_b))^2)/n
}
mean(boots) - variance # estimated bias
((n-1)/n)*1 -1 #true
修偏 =
方差的真实值1,naive variance = 0.95.
not working well on
但是真实值是1.
x = runif(100)
boots = rep(0,1000)
for(b in 1:1000){
boots[b] = max(sample(x,replace=TRUE))
}
quantile(boots,c(0.025,0.975))
Bootstrapping a single statistic
rsq <- function(formula , data , indices) {
d <- data[indices, ]
fit <- lm(formula, data = d)
return (summary(fit)$r.square)
}
library(boot)
set.seed(1234)
attach(mtcars)
results <- boot(data = mtcars, statistic = rsq, R = 1000, formula = mpg ~ wt + disp)
print(results)
plot(results)
boot.ci(results, type = c("perc", "bca"))
Bootstrapping several statistics
when adding or removing a few data points changes the estimator too much, the bootstrapping does not work well.
bs <- function(formula, data, indices){
d <- data[indices, ]
fit <- lm(formula, data = d)
return(coef(fit))
}
library(boot)
set.seed(1234)
results <- boot(data = mtcars, statistic = bs, R = 1000, formula = mpg ~ wt + disp)
print(results)
plot(results, index = 2)
print(boot.ci(results, type = "bca", index = 2))
print(boot.ci(results, type = "bca", index = 3))
Q:How large does the original sample need to be?
A:An original sample size of 20--30 is sufficient for good results, as long as the sample is representative of the population.
Q: How many replications are needed? A: I find that 1000 replications are more than adequate in most cases.



【推荐】国内首个AI IDE,深度理解中文开发场景,立即下载体验Trae
【推荐】编程新体验,更懂你的AI,立即体验豆包MarsCode编程助手
【推荐】抖音旗下AI助手豆包,你的智能百科全书,全免费不限次数
【推荐】轻量又高性能的 SSH 工具 IShell:AI 加持,快人一步
· Manus重磅发布:全球首款通用AI代理技术深度解析与实战指南
· 被坑几百块钱后,我竟然真的恢复了删除的微信聊天记录!
· 没有Manus邀请码?试试免邀请码的MGX或者开源的OpenManus吧
· 园子的第一款AI主题卫衣上架——"HELLO! HOW CAN I ASSIST YOU TODAY
· 【自荐】一款简洁、开源的在线白板工具 Drawnix