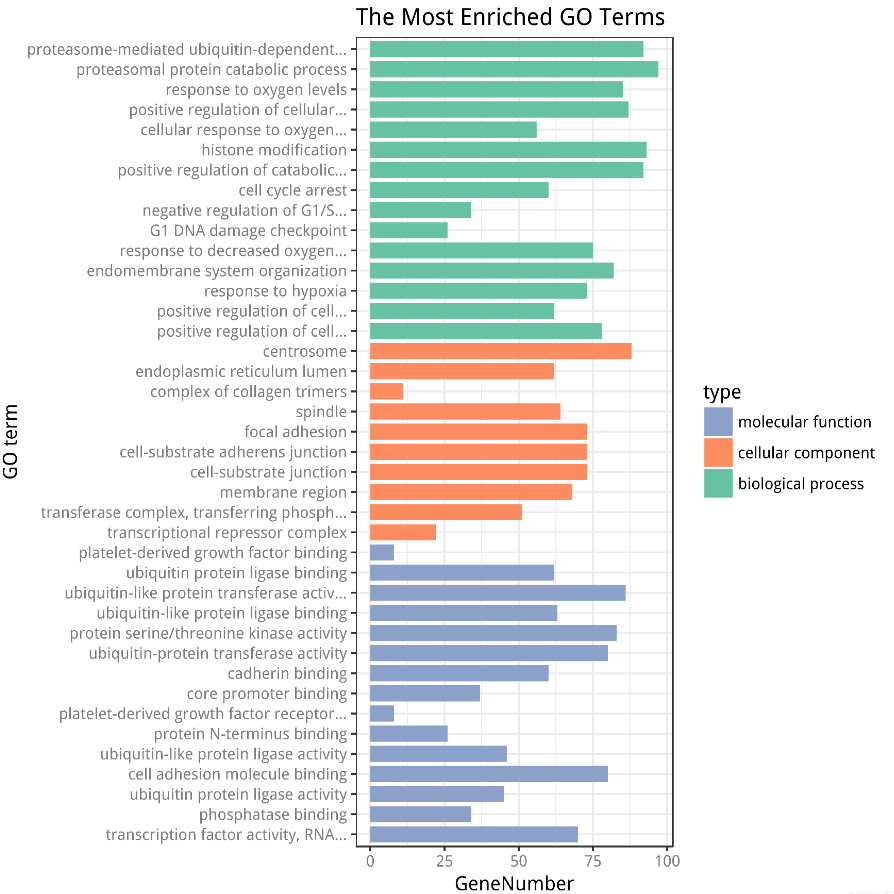
GO富集分析柱状图
1 target_gene_id <- unique(read.delim("miRNA-gene interactions.txt")$EntrezID) 2 # BiocInstaller::biocLite("clusterProfiler") 3 # BiocInstaller::biocLite("org.Hs.eg.db") 4 5 display_number = c(15, 10, 15) 6 ## GO enrichment with clusterProfiler 7 library(clusterProfiler) 8 ego_MF <- enrichGO(OrgDb="org.Hs.eg.db", 9 gene = target_gene_id, 10 pvalueCutoff = 0.05, 11 ont = "MF", 12 readable=TRUE) 13 ego_result_MF <- as.data.frame(ego_MF)[1:display_number[1], ] 14 # ego_result_MF <- ego_result_MF[order(ego_result_MF$Count),] 15 16 ego_CC <- enrichGO(OrgDb="org.Hs.eg.db", 17 gene = target_gene_id, 18 pvalueCutoff = 0.05, 19 ont = "CC", 20 readable=TRUE) 21 ego_result_CC <- as.data.frame(ego_CC)[1:display_number[2], ] 22 # ego_result_CC <- ego_result_CC[order(ego_result_CC$Count),] 23 24 ego_BP <- enrichGO(OrgDb="org.Hs.eg.db", 25 gene = target_gene_id, 26 pvalueCutoff = 0.05, 27 ont = "BP", 28 readable=TRUE) 29 ego_result_BP <- na.omit(as.data.frame(ego_BP)[1:display_number[3], ]) 30 # ego_result_BP <- ego_result_BP[order(ego_result_BP$Count),] 31 32 go_enrich_df <- data.frame(ID=c(ego_result_BP$ID, ego_result_CC$ID, ego_result_MF$ID), 33 Description=c(ego_result_BP$Description, ego_result_CC$Description, ego_result_MF$Description), 34 GeneNumber=c(ego_result_BP$Count, ego_result_CC$Count, ego_result_MF$Count), 35 type=factor(c(rep("biological process", display_number[1]), rep("cellular component", display_number[2]), 36 rep("molecular function", display_number[3])), levels=c("molecular function", "cellular component", "biological process"))) 37 38 ## numbers as data on x axis 39 go_enrich_df$number <- factor(rev(1:nrow(go_enrich_df))) 40 ## shorten the names of GO terms 41 shorten_names <- function(x, n_word=4, n_char=40){ 42 if (length(strsplit(x, " ")[[1]]) > n_word || (nchar(x) > 40)) 43 { 44 if (nchar(x) > 40) x <- substr(x, 1, 40) 45 x <- paste(paste(strsplit(x, " ")[[1]][1:min(length(strsplit(x," ")[[1]]), n_word)], 46 collapse=" "), "...", sep="") 47 return(x) 48 } 49 else 50 { 51 return(x) 52 } 53 } 54 55 labels=(sapply( 56 levels(go_enrich_df$Description)[as.numeric(go_enrich_df$Description)], 57 shorten_names)) 58 names(labels) = rev(1:nrow(go_enrich_df)) 59 60 ## colors for bar // green, blue, orange 61 CPCOLS <- c("#8DA1CB", "#FD8D62", "#66C3A5") 62 library(ggplot2) 63 p <- ggplot(data=go_enrich_df, aes(x=number, y=GeneNumber, fill=type)) + 64 geom_bar(stat="identity", width=0.8) + coord_flip() + 65 scale_fill_manual(values = CPCOLS) + theme_bw() + 66 scale_x_discrete(labels=labels) + 67 xlab("GO term") + 68 theme(axis.text=element_text(face = "bold", color="gray50")) + 69 labs(title = "The Most Enriched GO Terms") 70 71 p 72 73 pdf("go_enrichment_of_miRNA_targets.pdf") 74 p 75 dev.off() 76 77 svg("go_enrichment_of_miRNA_targets.svg") 78 p 79 dev.off()