聚类的三种实现方法
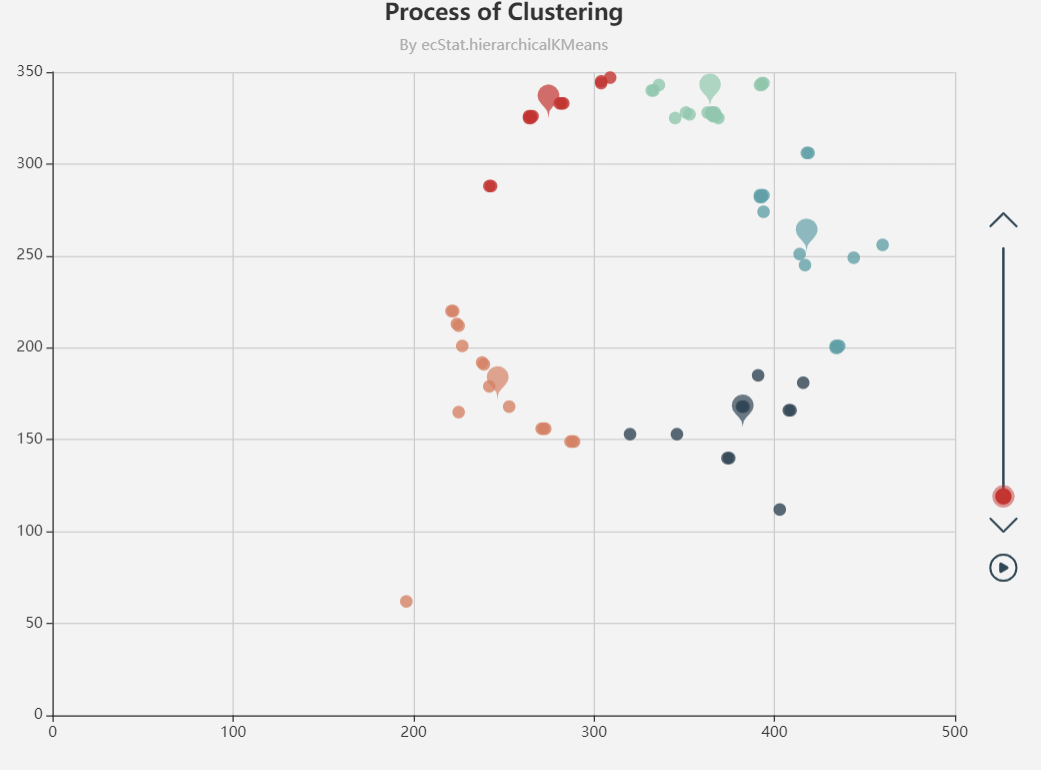
echarts聚类,还会画出质心,可视化效果挺好,适合网页程序应用。
https://www.echartsjs.com/examples/editor.html?c=scatter-clustering-process
var data = [
[196,62] ,
[221,220] ,
[222,220] ,
[224,213] ,
[225,165] ,
[225,212] ,
[227,201] ,
[238,192] ,
[239,191] ,
[242,179] ,
[242,288] ,
[243,288] ,
[253,168] ,
[264,325] ,
[264,326] ,
[265,325] ,
[265,326] ,
[266,326] ,
[271,156] ,
[272,156] ,
[273,156] ,
[281,333] ,
[282,333] ,
[283,333] ,
[287,149] ,
[288,149] ,
[289,149] ,
[304,344] ,
[304,345] ,
[309,347] ,
[320,153] ,
[332,340] ,
[333,340] ,
[336,343] ,
[345,325] ,
[346,153] ,
[351,328] ,
[353,327] ,
[363,328] ,
[365,327] ,
[365,328] ,
[366,326] ,
[366,327] ,
[366,328] ,
[367,326] ,
[367,327] ,
[367,328] ,
[368,326] ,
[369,325] ,
[374,140] ,
[375,140] ,
[382,168] ,
[383,168] ,
[391,185] ,
[392,282] ,
[392,283] ,
[392,343] ,
[393,282] ,
[393,283] ,
[393,343] ,
[393,344] ,
[394,274] ,
[394,283] ,
[394,344] ,
[403,112] ,
[408,166] ,
[409,166] ,
[414,251] ,
[416,181] ,
[417,245] ,
[418,306] ,
[419,306] ,
[434,200] ,
[434,201] ,
[435,200] ,
[435,201] ,
[436,201] ,
[444,249] ,
[460,256] ,
];
var clusterNumber = 5;
// See https://github.com/ecomfe/echarts-stat
var step = ecStat.clustering.hierarchicalKMeans(data, clusterNumber, true);
var result;
option = {
timeline: {
top: 'center',
right: 35,
height: 300,
width: 10,
inverse: true,
playInterval: 2500,
symbol: 'none',
orient: 'vertical',
axisType: 'category',
autoPlay: true,
label: {
normal: {
show: false
}
},
data: []
},
baseOption: {
title: {
text: 'Process of Clustering',
subtext: 'By ecStat.hierarchicalKMeans',
sublink: 'https://github.com/ecomfe/echarts-stat',
left: 'center'
},
xAxis: {
type: 'value'
},
yAxis: {
type: 'value'
},
series: [{
type: 'scatter'
}]
},
options: []
};
for (var i = 0; !(result = step.next()).isEnd; i++) {
option.options.push(getOption(result, clusterNumber));
option.timeline.data.push(i + '');
}
function getOption(result, k) {
var clusterAssment = result.clusterAssment;
var centroids = result.centroids;
var ptsInCluster = result.pointsInCluster;
var color = ['#c23531', '#2f4554', '#61a0a8', '#d48265', '#91c7ae', '#749f83', '#ca8622', '#bda29a', '#6e7074', '#546570', '#c4ccd3'];
var series = [];
for (i = 0; i < k; i++) {
series.push({
name: 'scatter' + i,
type: 'scatter',
animation: false,
data: ptsInCluster[i],
markPoint: {
symbolSize: 29,
label: {
normal: {
show: false
},
emphasis: {
show: true,
position: 'top',
formatter: function (params) {
return Math.round(params.data.coord[0] * 100) / 100 + ' ' +
Math.round(params.data.coord[1] * 100) / 100 + ' ';
},
textStyle: {
color: '#000'
}
}
},
itemStyle: {
normal: {
opacity: 0.7
}
},
data: [{
coord: centroids[i]
}]
}
});
}
return {
tooltip: {
trigger: 'axis',
axisPointer: {
type: 'cross'
}
},
series: series,
color: color
};
}
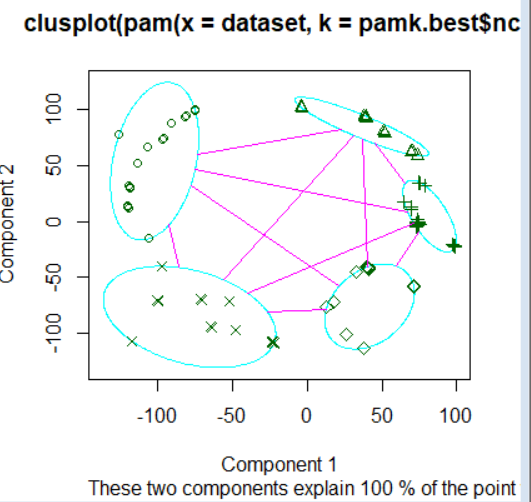
R语言聚类,具体参考https://www.cnblogs.com/think90/p/7133753.html
PAM(Partitioning Around Medoids) 围绕中心点的分割算法
k-means算法取得是均值,那么对于异常点其实对其的影响非常大,很可能这种孤立的点就聚为一类,一个改进的方法就是PAM算法,也叫k-medoids clustering
首先通过fpc包中的pamk函数得到最佳聚类数目,首先别忘了安装fpc包
install.packages("fpc")
library(fpc)
pamk.best <- pamk(dataset)
pamk.best$ncpamk函数不需要提供聚类数目,也会直接自动计算出最佳聚类数,这里也得到为3
得到聚类数提供给cluster包下的pam函数并进行可视化
library(cluster)
clusplot(pam(dataset, pamk.best$nc)) 
Java实现
参考https://blog.csdn.net/zuochao_2013/article/details/71423917
import java.io.File;
import java.io.IOException;
import java.util.ArrayList;
import java.util.List;
import org.apache.hadoop.conf.Configuration;
import org.apache.hadoop.fs.FileSystem;
import org.apache.hadoop.fs.Path;
import org.apache.hadoop.io.IntWritable;
import org.apache.hadoop.io.LongWritable;
import org.apache.hadoop.io.SequenceFile;
import org.apache.hadoop.io.Text;
import org.apache.mahout.clustering.WeightedVectorWritable;
import org.apache.mahout.clustering.kmeans.Cluster;
import org.apache.mahout.clustering.kmeans.KMeansDriver;
import org.apache.mahout.common.distance.EuclideanDistanceMeasure;
import org.apache.mahout.math.RandomAccessSparseVector;
import org.apache.mahout.math.Vector;
import org.apache.mahout.math.VectorWritable;
public class SimpleKMeansClustering {
public static final double[][] points = { {1, 1}, {2, 1}, {1, 2},
{2, 2}, {3, 3}, {8, 8},
{9, 8}, {8, 9}, {9, 9}};
public static void writePointsToFile(List<Vector> points,
String fileName,
FileSystem fs,
Configuration conf) throws IOException {
Path path = new Path(fileName);
SequenceFile.Writer writer = new SequenceFile.Writer(fs, conf,
path, LongWritable.class, VectorWritable.class);
long recNum = 0;
VectorWritable vec = new VectorWritable();
for (Vector point : points) {
vec.set(point);
writer.append(new LongWritable(recNum++), vec);
}
writer.close();
}
public static List<Vector> getPoints(double[][] raw) {
List<Vector> points = new ArrayList<Vector>();
for (int i = 0; i < raw.length; i++) {
double[] fr = raw[i];
Vector vec = new RandomAccessSparseVector(fr.length);
vec.assign(fr);
points.add(vec);
}
return points;
}
public static void main(String args[]) throws Exception {
int k = 2;
List<Vector> vectors = getPoints(points);
File testData = new File("testdata");
if (!testData.exists()) {
testData.mkdir();
}
testData = new File("testdata/points");
if (!testData.exists()) {
testData.mkdir();
}
Configuration conf = new Configuration();
FileSystem fs = FileSystem.get(conf);
writePointsToFile(vectors, "testdata/points/file1", fs, conf);
Path path = new Path("testdata/clusters/part-00000");
SequenceFile.Writer writer = new SequenceFile.Writer(fs, conf,
path, Text.class, Cluster.class);
for (int i = 0; i < k; i++) {
Vector vec = vectors.get(i);
Cluster cluster = new Cluster(vec, i, new EuclideanDistanceMeasure());
writer.append(new Text(cluster.getIdentifier()), cluster);
}
writer.close();
KMeansDriver.run(conf, new Path("testdata/points"), new Path("testdata/clusters"),
new Path("output"), new EuclideanDistanceMeasure(), 0.001, 10,
true, false);
SequenceFile.Reader reader = new SequenceFile.Reader(fs,
new Path("output/" + Cluster.CLUSTERED_POINTS_DIR
+ "/part-m-00000"), conf);
IntWritable key = new IntWritable();
WeightedVectorWritable value = new WeightedVectorWritable();
while (reader.next(key, value)) {
System.out.println(value.toString() + " belongs to cluster "
+ key.toString());
}
reader.close();
}
}




