背景
接触过一段时间遗传变异检测,现有公认的 GATK 检测小变异(snv + indel)的 “最佳实践” ,但是对于结构变异(SV),尽管有相当多软件能进行 SV 检测,但在其目前业内没有共识的检测软件和流程。现将自己的调研结果和成果记录下来。
SV 二代的检测原理主要有 4 个:
- RP : 基于双端测序数据,进行 SV 检测, 它不能精确到单个碱基断点。
- SR : 检测到的 SVs 断点能精确到单个碱基。
- RD : 基于 read 覆盖深度, 计算拷贝数变异。
- AS : 基于 de novo assembly。
此外还可能利用 SV 范围内 snv 变异辅助确定,比如二倍体 DUP 上的杂合变异 reads 的 ref/alt 倾向于 1/2 或 2/1, 而二倍体 DEL 范围内应该不存在杂合变异。各检测原理都有各自的优势,其中 AS 是相对最为准确的原理。文献综述推荐使用多种检测原理同时进行检测,最后综合各原理的检测结果进行筛选、过滤,以此提高检测灵敏度和准确度。
文献
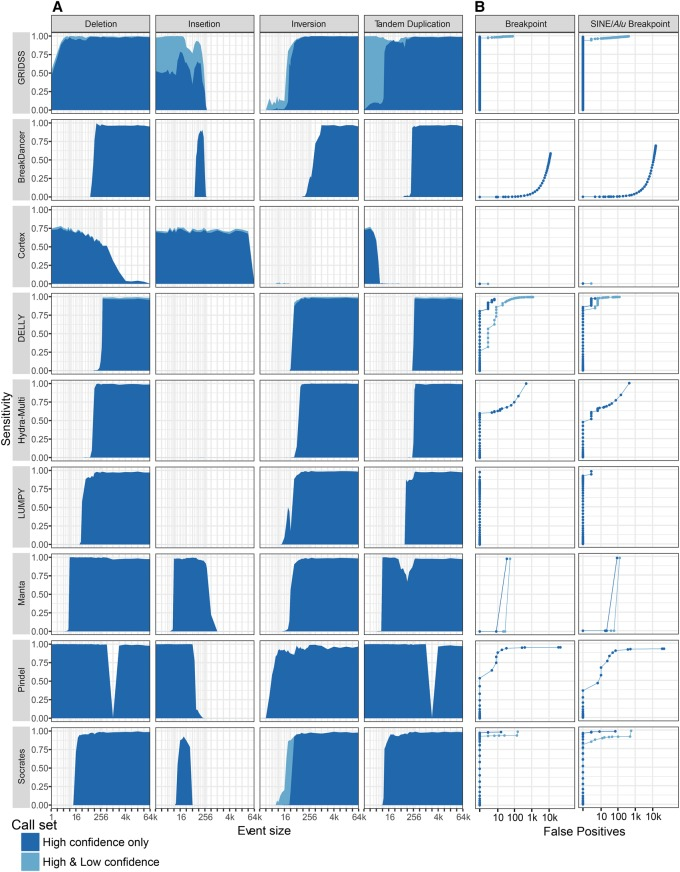
在 GRIDSS 文献选用了大家常用,比较可靠的 SV 检测软件进行比较全面的测试,从结果上看,多款软件都有各自的优势,如 gridss 和 lumpy 检测的断点位置最准确, manta、Socrates 检测的 SV 更全面,测试结果如下图:
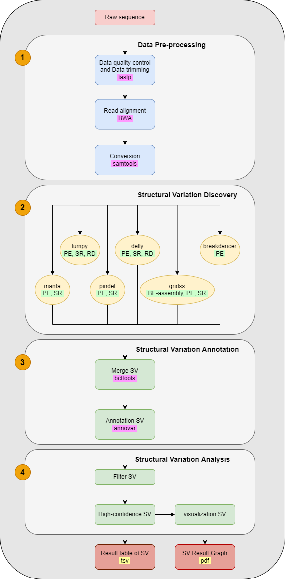
- 根据多个文献描述,预期 SV 检测流程图:
经验
SV 检测软件: lumpy,manta
SV vcf 合并:缺乏通用工具
自建 python 工具:
点击查看代码
import re
import os
import sys
import attr
import gzip
import logging
from typing import TextIO
from collections import defaultdict, OrderedDict
logging.basicConfig(
level = logging.INFO,
format = "[%(asctime)s - %(levelname)-4s - %(module)s : line %(lineno)s] %(message)s",
datefmt = "%Y-%m-%d %H:%M:%S"
)
@attr.s
class VCF_header:
""" vcf 表头储存类 """
SAMPLES = attr.ib(type=list, default=attr.Factory(list))
COL_HEARER = attr.ib(type=str, default='#CHROM\tPOS\tID\tREF\tALT\tQUAL\tFILTER\tINFO\tFORMAT')
def __attrs_post_init__(self):
self.REC_order = {
'fileformat': '',
'source': '',
'reference': '',
'contig': '',
'INFO': '',
'FORMAT': '',
'FILTER': '',
'ALT': '',
'OTHER': '',
}
def add(self, info: str):
for line in info.strip().split('\n'):
line = line.strip()
# 空行跳过
if not line:
continue
if line.startswith(f'##'):
key = re.findall(f'^##(\w+)=', line)[0]
if key in self.REC_order:
# vcf 文件类型只选用其中一个
if key == 'fileformat' and self.REC_order[key]:
continue
# 重复记录去除
if line in self.REC_order[key]:
continue
self.REC_order[key] = f"{self.REC_order[key]}{line}\n"
else:
self.REC_order['OTHER'] = f"{self.REC_order['OTHER']}{line}\n"
elif line.startswith(f'#CHROM'):
if not self.SAMPLES:
self.SAMPLES = line.split('\t')[9:]
# 增加表头时,核查样本是否一致
elif self.SAMPLES != line.split('\t')[9:]:
sys.exit(f'【Error】 样本编号不一致!')
else:
continue
# 增加新的 header 信息
add_info = [
['INFO', '##INFO=<ID=SVTOOL,Number=1,Type=String,Description="Tool used to generate the SV">'],
['INFO', '##INFO=<ID=NUM_SVTOOLS,Number=1,Type=Integer,Description="Number of SV callers supporting the event">'],
]
for k, v in add_info:
if k not in self.REC_order:
logging.warning(f'表头字段不存在 {k}, 忽略新增的表头: \n{v}')
continue
if v not in self.REC_order[k]:
logging.debug(f'新增 header 记录 {k} : {v}')
self.REC_order[k] = f"{self.REC_order[k]}{v}\n"
def merge(self, in_header):
""" 合并另一个 VCF_header 实例类 """
# 样本不一致时不合并
if self.SAMPLES != in_header.SAMPLES:
logging.error(f'样本不一致')
for v in in_header.REC_order.values():
self.add(v)
def __str__(self) -> str:
header_str = "".join([info for info in self.REC_order.values() if info])
all_sample = "\t".join(self.SAMPLES)
header_str = f"{header_str.strip()}\n{self.COL_HEARER}\t{all_sample}\n"
return header_str
@attr.s
class Recod:
""" vcf 单条记录储存类 """
CHROM = attr.ib(type=str, default=attr.Factory(str))
POS = attr.ib(type=str, default=attr.Factory(str))
ID = attr.ib(type=str, default=attr.Factory(str))
REF = attr.ib(type=str, default=attr.Factory(str))
ALT = attr.ib(type=str, default=attr.Factory(str))
QUAL = attr.ib(type=str, default=attr.Factory(str))
FILTER = attr.ib(type=str, default=attr.Factory(str))
INFO = attr.ib(type=defaultdict, default=attr.Factory(defaultdict))
FORMAT = attr.ib(type=list, default=attr.Factory(list))
SAMPLE_value = attr.ib(type=OrderedDict, default=attr.Factory(OrderedDict))
def __str__(self) -> str:
res = f"{self.CHROM}\t{self.POS}\t{self.ID}\t{self.REF}\t{self.ALT}\t{self.QUAL}\t{self.FILTER}\t"
for k, v in self.INFO.items():
res = f"{res}{k}={v};"
else:
res = res.strip(';')
res = f"{res}\t{':'.join(self.FORMAT)}\t"
for vs in self.SAMPLE_value.values():
for v in vs.values():
res = f"{res}{v}:"
else:
res = res.strip(':')
return res
@attr.s
class VCF_recods:
""" vcf 记录储存类,自动核查是否按染色体排序 """
SAMPLES = attr.ib(type=list, default=attr.Factory(list))
all_records = attr.ib(type=list, default=attr.Factory(list))
allow_chrom = attr.ib(type=list, default= [
str(i) for i in range(1,23)
] + [
f'CHR{i}' for i in range(1,23)
] + ['X', 'Y', 'CHRX', 'CHRY']
)
# 用于核对 add 添加的 record 是否按指定的染色体顺序
__last_chrom = attr.ib(type=str, default=attr.Factory(str))
__last_pos = attr.ib(type=str, default=attr.Factory(str))
def Iter(self) -> Recod:
""" 记录访问的生成器 """
for rec in self.all_records:
yield rec
def __str__(self) -> str:
return "\n".join([r.__str__() for r in self.all_records])
def add(self, info: str, svtool: str='', min_sv: int=50):
""" 添加新的记录 """
cells = info.strip().split('\t')
# print('cells', cells)
# 空行跳过,列数小于 10 (至少一个样本) 报错退出。
if not cells:
return
if len(cells) < 10:
logging.error(f'vcf 记录小于 10 列: \n{info}')
sys.exit('vcf 记录小于 10 列')
# 染色体合法性
cells[0] = cells[0].upper()
if cells[0] not in self.allow_chrom:
logging.debug(f'染色体不合法: \n{info}')
return
if 'SVTYPE=BND' in cells[7]:
alt_chr = re.findall(r'[\[\]]([\w\.]+):\d+', cells[4])[0]
if alt_chr not in self.allow_chrom:
logging.debug(f'alt 染色体不合法: \n{info}')
return
# 排序检查
if not self.__last_chrom:
self.__last_chrom = cells[0]
self.__last_pos = cells[1]
elif self.allow_chrom.index(cells[0]) < self.allow_chrom.index(self.__last_chrom) or \
(cells[0] == self.__last_chrom and int(self.__last_pos) > int(cells[1])):
# print('self.__last_chrom', self.__last_chrom)
# print('self.__last_pos', self.__last_pos)
logging.error(f'vcf 没有排序: {info}')
sys.exit()
record = Recod()
record.CHROM = cells[0]
record.POS = cells[1]
record.ID = cells[2]
record.REF = cells[3]
record.ALT = cells[4]
record.QUAL = cells[5]
record.FILTER = cells[6]
for val in cells[7].split(';'):
if '=' not in val:
record.INFO.update({val:val})
else:
k, v = val.split('=')
record.INFO.update({k:v})
# INFO 内添加 SVTOOL,NUM_SVTOOLS 字段
if 'NUM_SVTOOLS' not in record.INFO:
record.INFO['NUM_SVTOOLS'] = 1
if 'SVTOOL' not in record.INFO:
if svtool:
record.INFO['SVTOOL'] = svtool
else:
record.INFO['SVTOOL'] = 'unknown'
record.FORMAT = cells[8].split(':')
# 必须要有样本
if not self.SAMPLES:
logging.error(f'没有样本名,不能添加数据!: \n{info}')
return
# 核对样本数量是否一致
if len(self.SAMPLES) != len(cells) - 9:
logging.error(f'样本数量不一致!: \n{self.SAMPLES}\n{info}')
return
# SV 长度要达标
if 'SVLEN' in record.INFO:
if abs(int(record.INFO['SVLEN'])) < min_sv:
logging.debug(f'变异长度不达标, min sv len = {min_sv}bp')
return
# 添加样本的数据
for i, sample in enumerate(self.SAMPLES):
values = cells[9+i].split(':')
for j, k in enumerate(record.FORMAT):
if sample not in record.SAMPLE_value:
record.SAMPLE_value[sample] = OrderedDict()
try:
a = values[j]
except IndexError:
print(cells)
print(record.FORMAT)
print(values, j)
sys.exit()
if ',' in values[j]:
values[j] = values[j].split(',')[-1]
record.SAMPLE_value[sample][k] = values[j]
#* PR 字段与 PE 同义,增加 PE
if k == 'PR':
record.SAMPLE_value[sample]['PE'] = values[j]
# 'PR' 前面加上 'PE'
if 'PR' in record.FORMAT and 'PE' not in record.FORMAT:
record.FORMAT.insert(record.FORMAT.index('PR'), 'PE')
# 增加到记录的列表内
if record in self.all_records:
logging.warning(f'记录重复: {info}')
return
self.__last_chrom = cells[0]
self.__last_pos = cells[1]
self.all_records.append(record)
@attr.s
class VCF:
""""
将 vcf 数据存入 VCF 类对象, 特点:
a. 使用 ‘标签:vcf文件名’ 来标记 vcf 来源,如 GATK:germline.vcf.gz
b. 自动对输入 vcf 记录排序
vcf : 一个 vcf 文件,允许传入压缩格式,允许给 vcf 添加特殊来源标记。
如 GATK 来源的 vcf 文件 germline.vcf.gz, 可写成 GATK:germline.vcf.gz
"""
vcf = attr.ib(type=str, default=attr.Factory(str))
min_sv = attr.ib(type=int, default=attr.Factory(int))
def __attrs_post_init__(self):
""" 解析 vcf 文件,保存为 header, records 对象 """
self.svtool = None
if self.vcf.count(':') == 1:
self.svtool, self.vcf = self.vcf.split(':')
if not os.path.isfile(self.vcf):
logging.error(f'【error】 文件不存在 {self.vcf}')
sys.exit()
if not self.min_sv:
self.min_sv: int = 50
if self.vcf.endswith('.gz'):
Reader = gzip.open(self.vcf, 'rt')
else:
Reader = open(self.vcf)
# 确保 vcf 记录按顺序排列
Reader = self._sort_check(Reader)
self.header = VCF_header()
self.records = VCF_recods()
for line in Reader:
if line.startswith('#'):
self.header.add(line)
self.records.SAMPLES = self.header.SAMPLES
else:
self.records.add(line, self.svtool, self.min_sv)
Reader.close()
logging.info(f"<{self.svtool}> 变异数量: {len(self.records.all_records)}")
def __str__(self) -> str:
return self.header.__str__() + "\n".join([r.__str__() for r in self.records.all_records])
def merge(self, in_vcf, overlap_p=0.8):
""" 合并另一个 VCF_recods 实例类, 两记录相同类型变异重叠区域超过最大区域的 80% 就合并 """
# 没有信息或样本不一致,直接退出
if not in_vcf.header or not in_vcf.records or \
in_vcf.header.SAMPLES != self.header.SAMPLES:
logging.error(f'数据为空,或者样本不一致!')
return
# 合并 header
self.header.merge(in_vcf.header)
id = 1
new_all_recods = VCF_recods()
my_generator = self.Iter()
in_generator = in_vcf.Iter()
my_rec: Recod = next(my_generator, None)
in_rec: Recod = next(in_generator, None)
my_next = None
in_next = None
# couns = 0
while my_rec or in_rec:
# couns += 1
# if my_rec.POS == in_rec.POS== '152807507':
# if couns > 4000 and couns % 1000:
# print('my_rec', my_rec)
# print('in_rec', in_rec, end='\n\n')
# if couns > 5000:
# sys.exit()
# 输入项没有了时,直接保存原记录
if not in_rec:
my_rec.ID = id
id += 1
new_all_recods.all_records.append(my_rec)
my_rec = next(my_generator, None)
continue
# 仅有输入项,直接保存输入项
elif not my_rec:
in_rec.ID = id
id += 1
new_all_recods.all_records.append(in_rec)
in_rec = next(in_generator, None)
continue
# 处理一个 vcf 文件内存在多个位置相同的 SV
my_rec_list = []
in_rec_list = []
same_pos = False
while my_rec.CHROM == in_rec.CHROM and my_rec.POS == in_rec.POS:
if not my_next:
my_next = next(my_generator, None)
if my_next and my_next.CHROM == my_rec.CHROM and my_next.POS == my_rec.POS:
my_rec_list.append(my_rec)
my_rec = my_next
my_next = None
continue
in_next = next(in_generator, None)
if in_next and in_next.CHROM == in_rec.CHROM and in_next.POS == in_rec.POS:
in_rec_list.append(in_rec)
in_rec = in_next
continue
same_pos = True
break
my_rec_list.append(my_rec)
in_rec_list.append(in_rec)
done_rec = []
call_my = False # 判断是否需要载入下一个 my_rec
call_in = False # 判断是否需要载入下一个 in_rec
for my_rec in my_rec_list:
for in_rec in in_rec_list:
res = self._compare_records(my_rec, in_rec, id, overlap_p)
if res:
id += 1
new_all_recods.all_records.append(res)
done_rec.extend([my_rec, in_rec])
if not my_next:
call_my = True
if not in_next:
call_in = True
break
# 比较记录顺序,靠前的先写入 all_records 内
my_chr_index = self.records.allow_chrom.index(my_rec.CHROM)
in_chr_index = self.records.allow_chrom.index(in_rec.CHROM)
if my_chr_index < in_chr_index:
my_rec.ID = id
id += 1
new_all_recods.all_records.append(my_rec)
call_my = True
done_rec.append(my_rec)
continue
elif in_chr_index < my_chr_index:
in_rec.ID = id
id += 1
new_all_recods.all_records.append(in_rec)
call_in = True
done_rec.append(in_rec)
continue
else:
if int(my_rec.POS) < int(in_rec.POS):
my_rec.ID = id
id += 1
new_all_recods.all_records.append(my_rec)
call_my = True
done_rec.append(my_rec)
continue
elif int(in_rec.POS) < int(my_rec.POS):
in_rec.ID = id
id += 1
new_all_recods.all_records.append(in_rec)
call_in = True
done_rec.append(in_rec)
continue
# 位置相等后续一次性处理
for rec in my_rec_list + in_rec_list:
# 多个位置相等的记录比较完后,添加到结果表内,其他情况的记录留着后续再比较
if rec not in done_rec and same_pos:
rec.ID = id
id += 1
new_all_recods.all_records.append(rec)
same_pos = False
call_my = True
call_in = True
if my_next:
my_rec = my_next
my_next = None
else:
if call_my:
my_rec = next(my_generator, None)
call_my = False
if in_next:
in_rec = in_next
in_next = None
else:
if call_in:
in_rec = next(in_generator, None)
call_in = False
# if couns == 4000 and my_rec.POS == in_rec.POS== '24033178':
# print('next')
# print('my_next', my_next)
# print('in_next', in_next)
# print('my_rec', my_rec)
# print('in_rec', in_rec, end='\n\n')
logging.info(f'合并之后变异数量: {id-1}')
self.records = new_all_recods
def _compare_records(self, my_rec, in_rec, ID, overlap_p=0.8):
""" 比较两个位置是否可以合并,可以合并的话,返回一个新的 VCF_record 类,否则返回 None """
if my_rec.CHROM == in_rec.CHROM and \
my_rec.INFO['SVTYPE'] == in_rec.INFO['SVTYPE'] and \
abs(int(my_rec.POS) - int(in_rec.POS)) <= 5:
#* 断点
if my_rec.INFO['SVTYPE'] == 'BND':
my_alt_chr, my_alt_pos = re.findall(r'[\[\]](\w+)\:(\d+)', my_rec.ALT)[0]
in_alt_chr, in_alt_pos = re.findall(r'[\[\]](\w+)\:(\d+)', in_rec.ALT)[0]
# 另一端点染色体一致,断点位置位置偏差小于 5
if my_alt_chr == in_alt_chr and \
abs(int(my_alt_pos) - int(in_alt_pos)) <= 5 and \
abs(int(my_rec.POS) - int(in_rec.POS)) <= 5:
return self._merge_record(my_rec, in_rec, ID)
#* 常规变异有 SVLEN 或 END
elif my_rec.INFO['SVTYPE'] != 'INS' and 'END' in my_rec.INFO and 'END' in in_rec.INFO:
my_end = int(my_rec.INFO['END'])
in_end = int(in_rec.INFO['END'])
my_pos = int(my_rec.POS)
in_pos = int(in_rec.POS)
my_len = my_end - int(my_rec.POS)
in_len = in_end - int(in_rec.POS)
over_p = self.cal_overlap(my_pos, my_end, in_pos, in_end)
if over_p*100 >= overlap_p:
return self._merge_record(my_rec, in_rec, ID)
elif 'SVLEN' in my_rec.INFO and 'SVLEN' in in_rec.INFO:
my_len = abs(int(my_rec.INFO['SVLEN']))
in_len = abs(int(in_rec.INFO['SVLEN']))
over_p = in_len/my_len if my_len > in_len else my_len/in_len
if over_p*100 >= overlap_p:
return self._merge_record(my_rec, in_rec, ID)
@staticmethod
def cal_overlap(a: int, b: int, x: int, y: int) -> float:
"""
计算 2 个区间重叠百分比,返回 0-1
(a, b) 为一个区间, (x, y) 为一个区间
"""
if a > b:
a, b = b, a
if x > y:
x, y = y, x
ord = [a, b, x, y]
ord.sort()
# ab in xy
if a == ord[1] and b == ord[2]:
return (b-a)/(y-x)
# xy in ab
if x == ord[1] and y == ord[2]:
return (y-x)/(b-a)
# ab overlap xy
if a == ord[1] and y == ord[2]:
return (y-a)/(b-x)
if x == ord[1] and b == ord[2]:
return (b-x)/(y-a)
return 0.0
def _merge_record(self, my_rec, in_rec, ID: str):
""" 整理需要合并的 """
# 合并记录
new_record = Recod()
new_record.CHROM = my_rec.CHROM
new_record.ID = ID
# 偏差越小位置越准确
if my_rec.POS == in_rec.POS:
new_record.POS = my_rec.POS
elif 'CIPOS' in my_rec.INFO:
if 'CIPOS' in in_rec.INFO:
my_CIPOS = sum(abs(int(i)) for i in my_rec.INFO['CIPOS'].split(','))
in_CIPOS = sum(abs(int(i)) for i in in_rec.INFO['CIPOS'].split(','))
if my_CIPOS < in_CIPOS:
new_record.POS = my_rec.POS
else:
new_record.POS = in_rec.POS
else:
new_record.POS = my_rec.POS
elif 'CIPOS' in in_rec.INFO:
new_record.POS = in_rec.POS
else:
logging.warning(f'CIPOS 不存在: \n{my_rec}\n{in_rec}')
new_record.POS = my_rec.POS
# 偏差越小位置越准确
if 'CIEND' in my_rec.INFO:
if 'CIEND' in in_rec.INFO:
my_CIEND = sum(abs(int(i)) for i in my_rec.INFO['CIEND'].split(','))
in_CIEND = sum(abs(int(i)) for i in in_rec.INFO['CIEND'].split(','))
if my_CIEND < in_CIEND:
new_record.ALT = my_rec.ALT
new_record.REF = my_rec.REF
else:
new_record.ALT = in_rec.ALT
new_record.REF = in_rec.REF
else:
new_record.ALT = my_rec.ALT
new_record.REF = my_rec.REF
else:
new_record.ALT = in_rec.ALT
new_record.REF = in_rec.REF
new_record.QUAL = self._merge_QUAL(my_rec.QUAL, in_rec.QUAL)
new_record.FILTER = self._merge_FILTER(my_rec.FILTER, in_rec.FILTER)
new_record.INFO = self._merge_INFO(my_rec.INFO, in_rec.INFO)
new_record.FORMAT = self._merge_FORMAT(my_rec.FORMAT, in_rec.FORMAT)
new_record.SAMPLE_value = self._merge_SAMPLE_value(new_record.FORMAT, my_rec, in_rec)
#! 修正明显的错误
if new_record.INFO['SVTYPE'] != 'INS' and \
'SVLEN' in new_record.INFO and \
'END' in new_record.INFO:
if abs(int(new_record.INFO['SVLEN'])) != int(new_record.INFO['END']) - int(new_record.POS):
logging.warning(f'发现 SVLEN 异常,进行修复: [{new_record.CHROM}:{new_record.POS}]')
new_record.INFO['SVLEN'] = str(int(new_record.INFO['END']) - int(new_record.POS))
# DEL 的长度为负值
if new_record.INFO['SVTYPE'] == 'DEL':
new_record.INFO['SVLEN'] = f"-{new_record.INFO['SVLEN']}"
return new_record
@staticmethod
def _merge_QUAL(my_QUAL: str, in_QUAL: str) -> str:
""" 合并 QUAL 字段 """
if my_QUAL.isdigit():
if in_QUAL.isdigit():
if int(my_QUAL) > int(in_QUAL):
return my_QUAL
return in_QUAL
return my_QUAL
return in_QUAL
@staticmethod
def _merge_FILTER(my_FILTER, in_FILTER):
""" 合并 FILTER 字段 """
if my_FILTER != '.':
if in_FILTER != '.' and my_FILTER != in_FILTER:
return f"{my_FILTER},{in_FILTER}"
return my_FILTER
return in_FILTER
@staticmethod
def _merge_INFO(my_INFO: defaultdict, in_INFO: defaultdict) -> defaultdict:
""" 合并 INFO 字段 """
new_INFO = defaultdict()
# 合并必须 SV 类型一致
new_INFO['SVTYPE'] = my_INFO['SVTYPE']
# 不精确变异标识符
if 'IMPRECISE' in my_INFO:
if 'IMPRECISE' in in_INFO:
new_INFO['IMPRECISE'] = 'IMPRECISE'
# CIPOS
if 'CIPOS' in my_INFO:
if 'CIPOS' in in_INFO:
my_CIPOS = sum(abs(int(i)) for i in my_INFO['CIPOS'].split(','))
in_CIPOS = sum(abs(int(i)) for i in in_INFO['CIPOS'].split(','))
if my_CIPOS < in_CIPOS:
new_INFO['CIPOS'] = my_INFO['CIPOS']
else:
new_INFO['CIPOS'] = in_INFO['CIPOS']
else:
new_INFO['CIPOS'] = my_INFO['CIPOS']
elif 'CIPOS' in in_INFO:
new_INFO['CIPOS'] = in_INFO['CIPOS']
# END, CIEND
first = True
if 'CIEND' in my_INFO:
if 'CIEND' in in_INFO:
my_CIEND = sum(abs(int(i)) for i in my_INFO['CIEND'].split(','))
in_CIEND = sum(abs(int(i)) for i in in_INFO['CIEND'].split(','))
if my_CIEND < in_CIEND:
first = True
else:
first = False
else:
first = True
elif 'CIEND' in in_INFO:
first = False
if first:
if 'CIEND' in my_INFO:
new_INFO['CIEND'] = my_INFO['CIEND']
elif 'CIEND' in in_INFO:
new_INFO['CIEND'] = in_INFO['CIEND']
if 'END' in my_INFO:
new_INFO['END'] = my_INFO['END']
elif 'END' in in_INFO:
new_INFO['END'] = in_INFO['END']
# else:
# pass
# logging.warning(f'没有 END')
if 'SVLEN' in my_INFO:
new_INFO['SVLEN'] = my_INFO['SVLEN']
elif 'SVLEN' in in_INFO:
new_INFO['SVLEN'] = in_INFO['SVLEN']
# else:
# pass
# logging.warning(f'没有 SVLEN')
else:
if 'CIEND' in in_INFO:
new_INFO['CIEND'] = in_INFO['CIEND']
elif 'CIEND' in my_INFO:
new_INFO['CIEND'] = my_INFO['CIEND']
if 'END' in in_INFO:
new_INFO['END'] = in_INFO['END']
elif 'END' in my_INFO:
new_INFO['END'] = my_INFO['END']
# else:
# pass
# logging.warning(f'没有 END')
if 'SVLEN' in in_INFO:
new_INFO['SVLEN'] = in_INFO['SVLEN']
elif 'SVLEN' in my_INFO:
new_INFO['SVLEN'] = my_INFO['SVLEN']
# else:
# pass
# logging.warning(f'没有 SVLEN')
# SVTOOL NUM_SVTOOLS
if 'SVTOOL' in my_INFO:
if 'SVTOOL' in in_INFO:
new_INFO['SVTOOL'] = f"{my_INFO['SVTOOL']},{in_INFO['SVTOOL']}"
# new_INFO['SVTOOL'] = ",".join(list(set(my_INFO['SVTOOL'].split(',') + in_INFO['SVTOOL'].split(','))))
new_INFO['NUM_SVTOOLS'] = str(new_INFO['SVTOOL'].count(',') + 1)
else:
logging.error(f'INFO 内 SVTOOL 字段丢失: {in_INFO}')
else:
logging.error(f'INFO 内 SVTOOL 字段丢失: {my_INFO}')
if 'NUM_SVTOOLS' in my_INFO:
if 'NUM_SVTOOLS' in in_INFO:
new_INFO['NUM_SVTOOLS'] = str(int(my_INFO['NUM_SVTOOLS']) + int(in_INFO['NUM_SVTOOLS']))
else:
logging.error(f'INFO 内 NUM_SVTOOLS 字段丢失: {in_INFO}')
else:
logging.error(f'INFO 内 NUM_SVTOOLS 字段丢失: {in_INFO}')
# other
for key in ['BND_DEPTH', 'MATE_BND_DEPTH']:
if key in my_INFO:
if key in in_INFO:
my_value = int(my_INFO[key])
in_value = int(in_INFO[key])
if my_value < in_value:
new_INFO[key] = my_INFO[key]
else:
new_INFO[key] = in_INFO[key]
else:
new_INFO[key] = my_INFO[key]
elif key in in_INFO:
new_INFO[key] = in_INFO[key]
return new_INFO
@staticmethod
def _merge_FORMAT(my_FORMAT: list, in_FORMAT: list) -> list:
""" 合并 样本信息 字段 """
allow_FORMAT = ['GT', 'PE', 'SR']
new_FORMAT = []
for key in my_FORMAT + in_FORMAT:
if key in allow_FORMAT and key not in new_FORMAT:
new_FORMAT.append(key)
return new_FORMAT
@staticmethod
def _merge_SAMPLE_value(FORMAT, my_rec, in_rec) -> OrderedDict:
""" 合并 样本信息 字段 """
new_SAMPLE_value = OrderedDict()
# PR 当作 PE 的别名
my_format = my_rec.FORMAT
in_format = in_rec.FORMAT
for s, _ in my_rec.SAMPLE_value.items():
new_SAMPLE_value[s] = OrderedDict()
for key in FORMAT:
if key == 'GT':
my_gt = my_rec.SAMPLE_value[s][key]
in_gt = in_rec.SAMPLE_value[s][key]
if my_gt != './.':
new_SAMPLE_value[s][key] = my_gt
elif in_gt != './.':
new_SAMPLE_value[s][key] = in_gt
else:
new_SAMPLE_value[s][key] = './.'
continue
if key in my_format:
my_v = my_rec.SAMPLE_value[s][key]
if key in in_format:
in_v = in_rec.SAMPLE_value[s][key]
if int(my_v) > int(in_v):
new_SAMPLE_value[s][key] = my_v
else:
new_SAMPLE_value[s][key] = in_v
else:
new_SAMPLE_value[s][key] = my_v
elif key in in_format:
in_v = in_rec.SAMPLE_value[s][key]
new_SAMPLE_value[s][key] = in_v
else:
logging.error(f'哪里有问题,两个 record 内都没有的话 {key} 哪来的丫!')
return new_SAMPLE_value
def Iter(self) -> Recod:
""" 记录访问的生成器 """
for rec in self.records.all_records:
yield rec
def write(self, out_file: str='out.vcf'):
if out_file.endswith('vcf.gz'):
o_vcf = gzip.open(out_file, 'wt')
else:
o_vcf = open(out_file, 'w')
o_vcf.write(self.header.__str__() + "\n".join([r.__str__() for r in self.records.all_records]))
@staticmethod
def _sort_check(in_reader: TextIO):
""" 对 vcf 记录按染色体编号进行排序 """
all_records = defaultdict(list)
for line in in_reader:
if line.startswith('#'):
yield line
else:
chrom, pos, *_ = line.split('\t')
chrom = chrom.upper().replace('CHR', '')
# 为了保证顺序符合预期,小数位的染色体前面填充 0
if chrom.isdigit():
key = "{:0>2d}_{:0>9d}".format(int(chrom), int(pos))
else:
key = "{:s}_{:0>9d}".format(chrom, int(pos))
all_records[key].append(line)
all_key = list(all_records.keys())
all_key.sort()
for key in all_key:
for info in all_records[key]:
yield info
if __name__ == '__main__':
in_vcf = VCF('lumpy:lumpy.vcf')
ou_vcf = VCF('manta:manta.vcf.gz')
in_vcf.merge(ou_vcf)
in_vcf.write()
SV 注释: AnnotSV
SV 可视化: samplot
注意事项
- SV 的假阳性结果特别多,需要根据一定的条件进行过滤
- 断点明确的 SV 可信度更高

 浙公网安备 33010602011771号
浙公网安备 33010602011771号