RGAugury鉴定RLP+RLK相关抗性基因
文献
Li, P., Quan, X., Jia, G. et al. RGAugury: a pipeline for genome-wide prediction of resistance gene analogs (RGAs) in plants. BMC Genomics 17, 852 (2016). https://doi.org/10.1186/s12864-016-3197-x
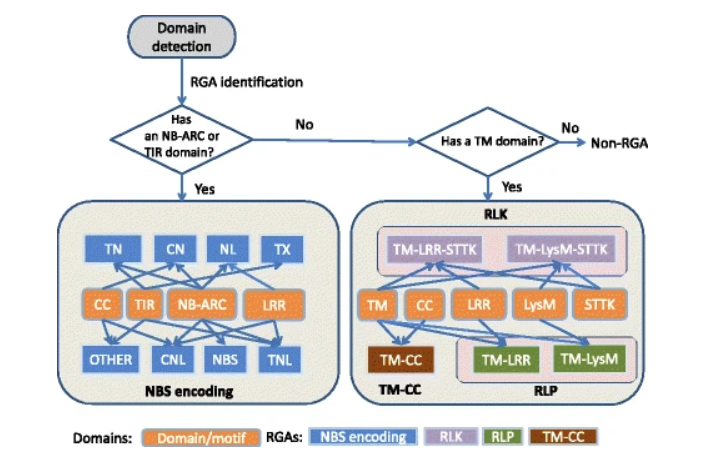
原理(RGA identification)
官网
报错相关
https://www.jianshu.com/p/358e5522ef57
gff文件输入注意处理
https://www.jianshu.com/p/358e5522ef57
尝试docker开搞
一、数据库本地化
pfam准备
mkdir ~/database/pfam
wget https://ftp.ebi.ac.uk/pub/databases/Pfam/current_release/Pfam-A.hmm.gz
wget https://ftp.ebi.ac.uk/pub/databases/Pfam/current_release/Pfam-A.hmm.dat.gz
wget http://ftp.ebi.ac.uk/pub/databases/Pfam/current_release/active_site.dat.gz
pigz -d Pfam-A.hmm.gz
pigz -d Pfam-A.hmm.dat.gz
hmmpress Pfam-A.hmminterproscan准备
wget https://ftp.ebi.ac.uk/pub/software/unix/iprscan/5/5.62-94.0/interproscan-5.62-94.0-64-bit.tar.gz
wget https://ftp.ebi.ac.uk/pub/software/unix/iprscan/5/5.62-94.0/interproscan-5.62-94.0-64-bit.tar.gz.md5
md5sum -c interproscan-5.62-94.0-64-bit.tar.gz.md5
tar -pxvzf interproscan-5.62-94.0-*-bit.tar.gz
cd interproscan-5.62-94.0
python3 setup.py -f interproscan.properties二、docker拉镜像
docker image pull yaanlpc/rgaugury:2.2三、启动docker
注意修改source路径到对应下载好pfam和interproscan目录,input目录放用于输入的pep、gff、genome文件。(将数据库、输入数据链接至容器内运行)
docker run -it \
--mount type=bind,source=/home/pingchuan/docker_project/database/pfam,target=/opt/pfam \
--mount type=bind,source=/home/pingchuan/docker_project/database/interproscan,target=/opt/interproscan \
--mount type=bind,source=/home/pingchuan/docker_project/input,target=/root/input \
yaanlpc/rgaugury:2.2 /bin/bash \四、正式使用软件
perl -S RGAugury.pl -p sample.fas -c 2 -pfx test
五、注意事项
Pfam文件找不到->检查hmmpress Pfam-A.hmm
interproscan相关报错->vim interproscan.properties文件关闭网络查找选择使用本地,对应precalculated.match.lookup.service.url=https://www.ebi.ac.uk/interpro/match-lookup置为空即可
Cvit可视化模块出错Cant open img->检查gff文件以及输入序列是否符合格式(似乎是需要带序列的gff)或者换用别的软件可视化
docker相关报错“invalid argument type=bind” -> type后面那一串不要加额外的空格分割否则参数读取错误
docker相关报错“permission denied” -> docker命令前加sudo


 浙公网安备 33010602011771号
浙公网安备 33010602011771号