WGDI学习记录之实战分析(二)
References
数据来源:https://banana-genome-hub.southgreen.fr/organism/1?tripal_pane=group_data
官方文档:https://wgdi.readthedocs.io/en/latest/Introduction.html
b站视频1: https://www.bilibili.com/video/BV1qK4y1U7eK/?spm_id_from=333.337.search-card.all.click&vd_source=934e01f1fb91e6f31db7ee191d595870
b站视频2: https://www.bilibili.com/video/BV195411P7L1/?spm_id_from=333.337.search-card.all.click
参考教程1:https://blog.csdn.net/u012110870/article/details/113544271
参考教程2: https://blog.csdn.net/m0_49960764/article/details/120141944?spm=1001.2101.3001.6650.5&utm_medium=distribute.pc_relevant.none-task-blog-2%7Edefault%7EBlogCommendFromBaidu%7ERate-5-120141944-blog-113544271.pc_relevant_3mothn_strategy_recovery&depth_1-utm_source=distribute.pc_relevant.none-task-blog-2%7Edefault%7EBlogCommendFromBaidu%7ERate-5-120141944-blog-113544271.pc_relevant_3mothn_strategy_recovery&utm_relevant_index=6
STEP00 准备数据
###下载数据
wget https://banana-genome-hub.southgreen.fr/filebrowser/download/524
wget https://banana-genome-hub.southgreen.fr/filebrowser/download/520
wget https://banana-genome-hub.southgreen.fr/filebrowser/download/521
mv 524 mac.gff
mv 520 mac.cds.fa
mv 521 mac.pep.fa###格式化数据
python 01.getgff.py mac.gff old.gff
python 02.gff_lens.py old.gff mac1s.gff mac1s.lens
python 03.seq_newname.py mac1s.gff mac.cds.fa mac1s.cds.fa
python 03.seq_newname.py mac1s.gff mac.pep.fa mac1s.pep.fa###这里的gff/cds/pep文件里包含了mito信息,此处分析不需要故删除,处理完后进行后续分析
###后续的分析主要是基于基因的信息
###这里先获取blast文件(wgdi作者推荐使用blastp)
makeblastdb -in mac1s.pep.fa -dbtype prot
blastp -num_threads 48 -db mac1s.pep.fa -query mac1s.pep.fa -outfmt 6 -evalue 1e-5 -num_alignments 20 -out mac1s_mac1s.blast
STEP01 dotplot
###有了blast文件,先画个散点图看看共线性吧(这里工具就比较多了,像JCVI中也可以做共线性散点图)
wgdi -d ? >> all.conf
*************
[dotplot]
blast = mac1s_mac1s/mac1s_mac1s.blast
gff1 = mac1s.gff
gff2 = mac1s.gff
lens1 = mac1s.lens
lens2 = mac1s.lens
genome1_name = M.acuminata
genome2_name = M.acuminata
multiple = 1
score = 100
evalue = 1e-5
repeat_number = 20
position = order
blast_reverse = false
ancestor_left = none
ancestor_top = none
markersize = 0.5
figsize = 10,10
savefig = 01mac_mac.dotplot.order.png
****************
wgdi -d all.conf 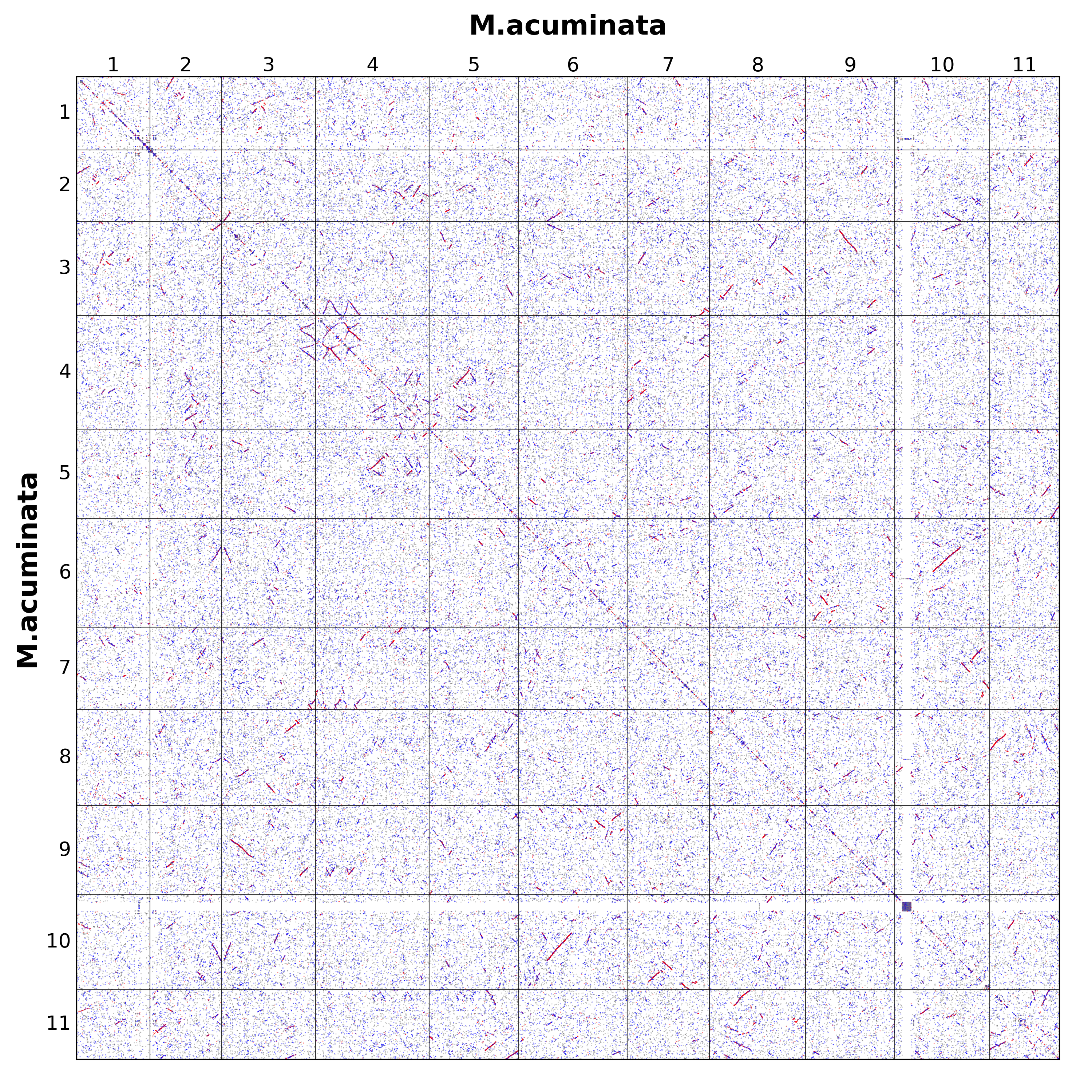
###从图中,我们很容易地就能观察到三种颜色,分别是红色,蓝色,和灰色。在WGDI中,红色表示genome2的基因在genome1中的最优同源(相似度最高)的匹配,次好的四个基因是蓝色,而剩余部分是为灰色。图中对角线出现的片段仅仅是因为仍存在一些串联重复基因而已。
###如果基因组存在加倍事件,那么对于一个基因组的某个区域可能会存在另一个区域与其有着相似的基因(同源基因),并且这些基因的排布顺序较为一致。反应到点阵图上,就是能够观察到有多个点所组成的"线"。这里的线需要有一个引号,因为当你放大之后,你会发现其实还是点而已。
###由于进一步鉴定共线性区域就建立在这些"点"的基础上,那么影响这些点是否为同源基因的参数就非常重要了,即配置文件中的score,evalue,repeat_number。
STEP02 collinearity+ks
###初步查看了共线性的散点图后,通过collinearity步骤提取共线性基因对儿信息
wgdi -icl ? >> all.conf
***************
[collinearity]
gff1 = mac1s.gff
gff2 = mac1s.gff
lens1 = mac1s.lens
lens2 = mac1s.lens
blast = mac1s_mac1s/mac1s_mac1s.blast
blast_reverse = false
multiple = 1
process = 8
evalue = 1e-5
score = 100
grading = 50,40,25
mg = 40,40
pvalue = 0.2
repeat_number = 10
positon = order
savefile = mac1s_mac1s.collinearity
***************
wgdi -icl all.conf###计算ks
wgdi -ks ? >> all.conf
***************
[ks]
cds_file = mac1s.cds.fa
pep_file = mac1s.pep.fa
align_software = muscle
pairs_file = mac1s_mac1s.collinearity
ks_file = mac1s.ks
***************
wgdi -ks all.conf###使用-bi合并collinearity以及ks信息(其中collinearity和ks是前两步生成的输出文件,ks_col 是声明使用ks文件里的哪一列)
wgdi -bi ? >> all.conf
***********************
[blockinfo]
gff1 = mac1s.gff
gff2 = mac1s.gff
lens1 = mac1s.lens
lens2 = mac1s.lens
blast = mac1s_mac1s/mac1s_mac1s.blast
collinearity = mac1s_mac1s.collinearity
score = 100
evalue = 1e-5
repeat_number = 20
position = order
ks = mac1s.ks
ks_col = ks_NG86
savefile = mac1s_mac1s.blockinfo.csv
***********************
wgdi -bi all.conf###随后涉及csv输出结果的整理建议参考孙老师b站视频做处理。



