Circos作图进阶系列(一)
今天跟着NCB的这篇文章学习circos作图各个模块的配置方法。关于circos.conf部分稍做了修改,使各个模块独立,结构更加清晰。作图代码及数据均来自于作者Github。其中配置contig组成展示的部分值得学习。
### 作图代码:
https://github.com/institut-de-genomique/Pahang-associated-data/tree/main/Figure1
### 数据来源:
Belser C, Baurens FC, Noel B, Martin G, Cruaud C, Istace B, Yahiaoui N, Labadie K, Hřibová E, Doležel J, Lemainque A, Wincker P, D'Hont A, Aury JM. Telomere-to-telomere gapless chromosomes of banana using nanopore sequencing. Commun Biol. 2021 Sep 7;4(1):1047. doi: 10.1038/s42003-021-02559-3. PMID: 34493830; PMCID: PMC8423783.
https://github.com/institut-de-genomique/Pahang-associated-data/tree/main/Figure1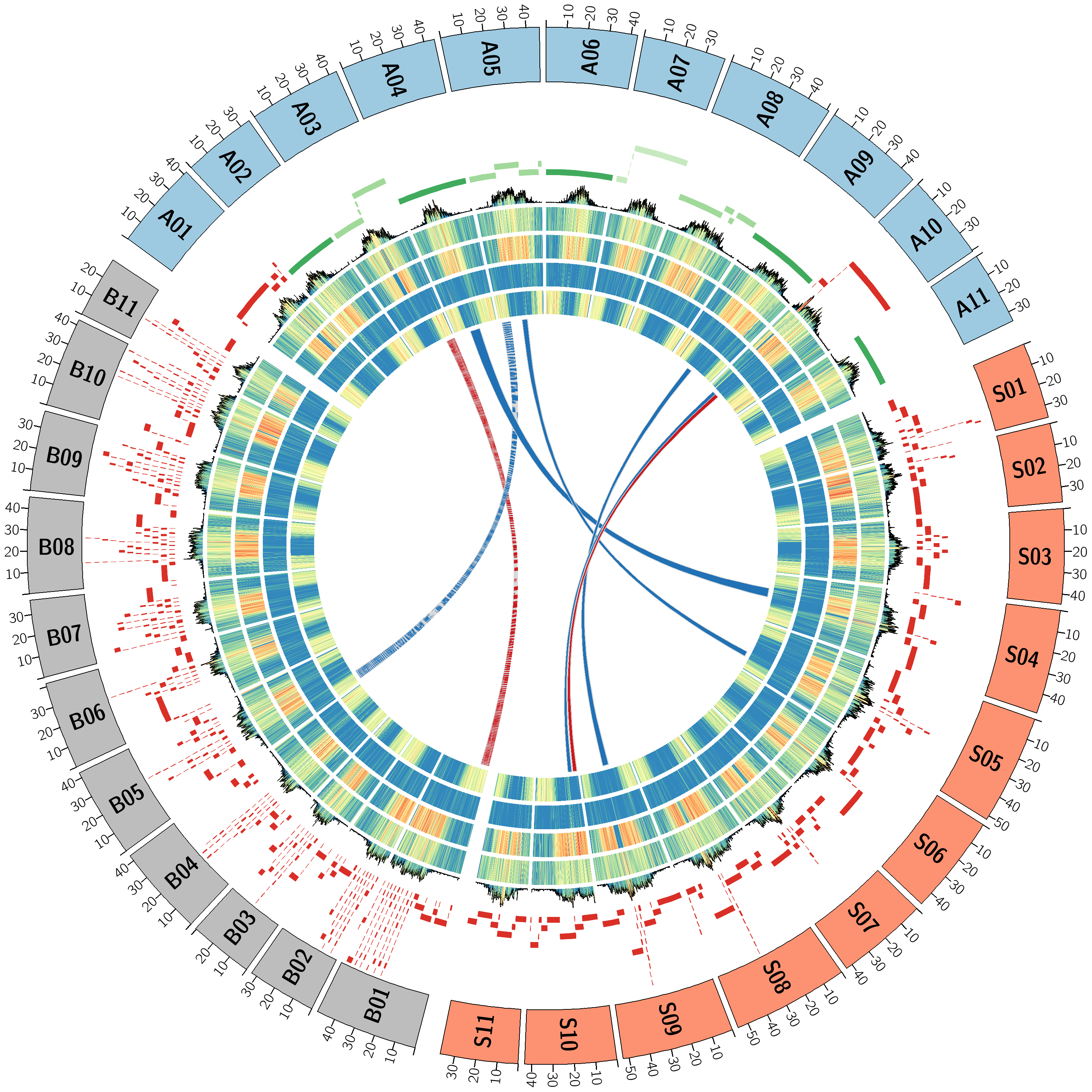
一、数据准备
作者提供的包里已经有了如下数据,作为circos的数据输入。以下输入数据来自于基因组重复注释gff文件、基因注释gff文件、共线性搜索等分析。
查看目录结构。
二、Circos配置
<Circos.conf> 主文件配置,接下来依次配置各个模块。最末两行为引用软件etc中已有模块,无需改动。
karyotype = karyotype.txt
chromosomes_units = 1000000
chromosomes_display_default = yes
<<include ideogram.conf>>
<<include ticks.conf>>
<<include image.conf>>
<<include plots.conf>>
<<include links.conf>>
<<include etc/colors_fonts_patterns.conf>>
<<include etc/housekeeping.conf>><ideogram.conf> 这个模块主要控制染色体排版方式,包括染色体间隔、填充颜色、染色体标签等。
<ideogram>
<spacing>
default = 3u
break = 1u
<pairwise A11 S01>
spacing = 12u
</pairwise>
<pairwise S11 B01>
spacing = 12u
</pairwise>
<pairwise B11 A01>
spacing = 12u
</pairwise>
</spacing>
# thickness (px) of chromosome ideogram
thickness = 150p
stroke_thickness = 2
# ideogram border color
stroke_color = black
fill = yes
# the default chromosome color is set here and any value
# defined in the karyotype file overrides it
fill_color = blue
# fractional radius position of chromosome ideogram within image
radius = 0.95r
show_label = yes
label_with_tag = yes
label_font = condensedbold
label_radius = dims(ideogram,radius) - 0.1r
label_size = 48
</ideogram><ticks.conf> 主要控制染色体上刻度显示。
show_ticks = yes
show_tick_labels = yes
<ticks>
skip_first_label = no
skip_last_label = no
radius = dims(ideogram,radius_outer)
min_label_distance_to_edge = 10p
label_offset = 2p
label_size = 8p
multiplier = 1e-6
color = black
<tick>
spacing = 10u
size = 20p
thickness = 3p
color = black
show_label = yes
label_size = 30p
label_offset = 0p
format = %d
</tick>
</ticks><image.conf> 该模块主要控制图像输出、设置图像半径、颜色背景、排版旋转角度等
<image>
svg = yes
dir = ./
file = circos-Musa_genomes_comparison.png
# radius of inscribed circle in image
radius = 1500p
background = white
# by default angle=0 is at 3 o'clock position
angle_offset = -90
</image><plots.conf> 该模块属于绘图模块,可以绘制histogram、dotplot、lineplot、heatmap等,均使用相同的数据输入格式。对应从外至内依次绘制展示6圈不同的数据。第二圈主要显示contig、其他的主要显示重复序列密度或者基因CDS密度。
<plots>
<plot>
type = tile
file = ALL_contigs.bed
r1 = 0.99r
r0 = 0.80r
layers = 12
thickness = 15
padding = 8
color = vdblue
stroke_color = vdblue
stroke_thickness = 1
show = yes
<rules>
<rule>
condition = var(id) ==1
stroke_color = dgreen
color = dgreen
flow = continue
</rule>
<rule>
condition = var(id) ==2
stroke_color = green
color = green
</rule>
<rule>
condition = var(id) ==3
stroke_color = green
color = green
</rule>
<rule>
condition = var(id) ==4
stroke_color = lgreen
color = lgreen
</rule>
<rule>
condition = var(id) == 5
stroke_color = vlgreen
color = vlgreen
</rule>
<rule>
condition = var(id) > 5
stroke_color = acen
color = acen
</rule>
</rules>
</plot>
<plot>
type = histogram
stroke_type = outline
thickness = 2
color = black
extend_bin = yes
file = ALL_centro.density.100kb
show = yes
r1 = 0.79r
r0 = 0.74r
<backgrounds>
<background>
color = white #vvlgrey
</background>
</backgrounds>
<rules>
<rule>
condition = 1
fill_color = eval(sprintf("spectral-9-div-%d",remap_int(var(value),30,100,9,1)))
</rule>
</rules>
</plot>
<plot>
type = heatmap
color = spectral-9-div-rev
file = ALL_gypsy.density.100kb
scale_log_base = 0.5
show = yes
r1 = 0.73r
r0 = 0.68r
</plot>
<plot>
type = heatmap
color = spectral-9-div-rev
file = ALL_copia.density.100kb
scale_log_base = 0.5
show = yes
r1 = 0.67r
r0 = 0.62r
</plot>
<plot>
type = heatmap
color = spectral-9-div-rev
file = ALL_dna.transposon.density.100kb
scale_log_base = 1
show = yes
r1 = 0.61r
r0 = 0.56r
</plot>
<plot>
type = heatmap
color = spectral-9-div-rev
file = ALL_genesCDS.density.100kb
scale_log_base = 1
show = yes
r1 = 0.55r
r0 = 0.50r
</plot>
</plots><links.conf> 该模块主要展示染色体间的共线性。通过对共线性块bed文件增加id列,然后通过rule增加condition匹配,来实现对不同的共线性块标记不同的颜色。
<links>
radius = 0.49r
crest = 1
ribbon = yes
flat = yes
bezier_radius = 0r
bezier_radius_purity = 0.5
<link>
file = ALL_syntenie.bed
stroke_color = vlgrey
stroke_thickness = 2
color = vlgrey
<rules>
<rule>
condition = var(id) ==1
color = vdred
flow = continue
</rule>
<rule>
condition = var(id) ==2
color = vdblue
flow = continue
</rule>
</rules>
</link>
</links>三、绘图
进入conf目录,使用circos一键绘图
circos -conf circos.conf运行成功后,当前目录下便会生成circos-Musa_genomes_comparison.svg以及circos-Musa_genomes_comparison.png



