JCVI作图进阶系列(二)
local synteny plot, focus on gene-level show matching regions along with aligned gene models.
参考自官网教程pairwise-synteny-plot
PS:第一步输入文件源自系列(一)的输出结果
一、准备工作
#Compute the layout for the gene-level matchings
python -m jcvi.compara.synteny mcscan grape.bed grape.peach.lifted.anchors --iter=1 -o grape.peach.i1.blocks#Choose any arbitrary region to visualize
head -50 grape.peach.i1.blocks > blocks#conbime *.bed(grape_peach.bed)
cat grape.bed peach.bed > grape_peach.bed#make blocks.layout(blocks.layout)
# x, y, rotation, ha, va, color, ratio, label
0.5, 0.6, 0, left, center, m, 1, grape Chr1
0.5, 0.4, 0, left, center, #fc8d62, 1, peach scaffold_1
# edges
e, 0, 1二、作图
使用jcvi.graphics.synteny模块,尝试不同作图风格,结果见blocks.pdf
#default_style
python -m jcvi.graphics.synteny blocks grape_peach.bed blocks.layout --notex
#line_shadestyle
python -m jcvi.graphics.synteny blocks grape_peach.bed blocks.layout --shadestyle=line --notex
#arrow_glyphstyle
python -m jcvi.graphics.synteny blocks grape_peach.bed blocks.layout --glyphstyle=arrow --notex
#orthogroup_glyphcolor(同源块颜色对应)
python -m jcvi.graphics.synteny blocks grape_peach.bed blocks.layout --glyphcolor=orthogroup --notex
#show all gene_labels
python -m jcvi.graphics.synteny blocks grape_peach.bed blocks.layout --genelabelsize 6 --notex
#show a few gene_labels
python -m jcvi.graphics.synteny blocks grape_peach.bed blocks.layout --genelabelsize=10 --genelabels=GSVIVT01012244001,Prupe.1G289800.1,GSVIVT01012225001,Prupe.1G288000.1 --notex三、结果
此处展示不同作图参数作图的结果blocks.pdf
#blocks1.pdf(default)
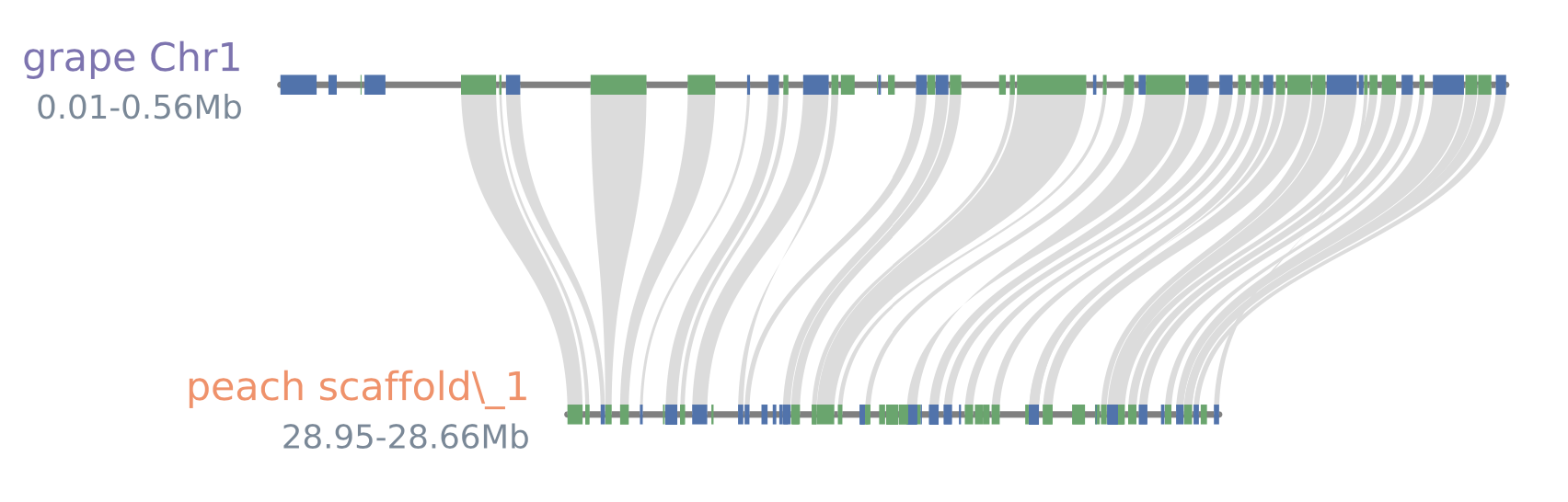
#blocks2.pdf(--shadestyle=line)
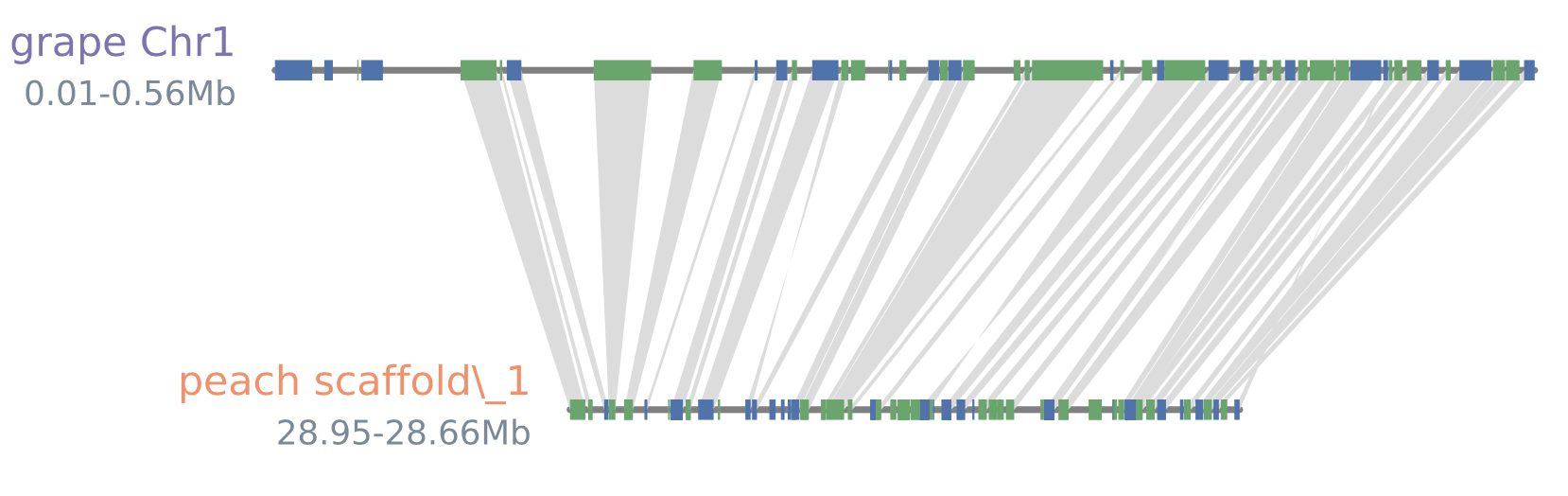
#blocks3.pdf(--glyphstyle=arrow)
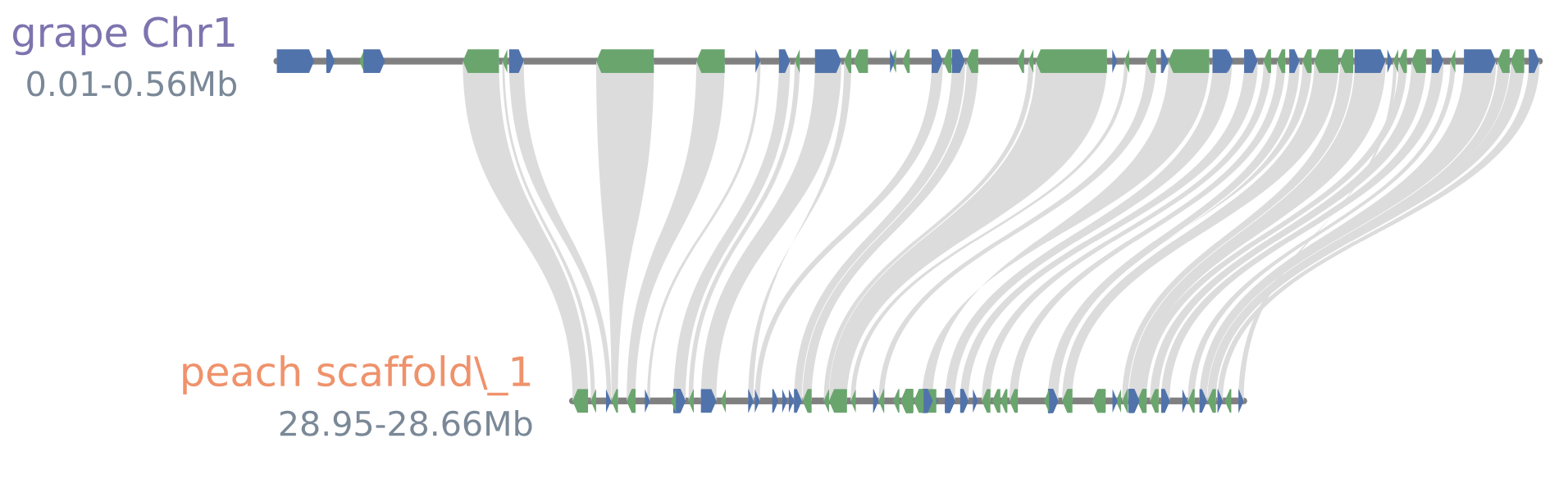
#blocks4.pdf(--glyphcolor=orthogroup)

#blocks5.pdf(--genelabelsize 6)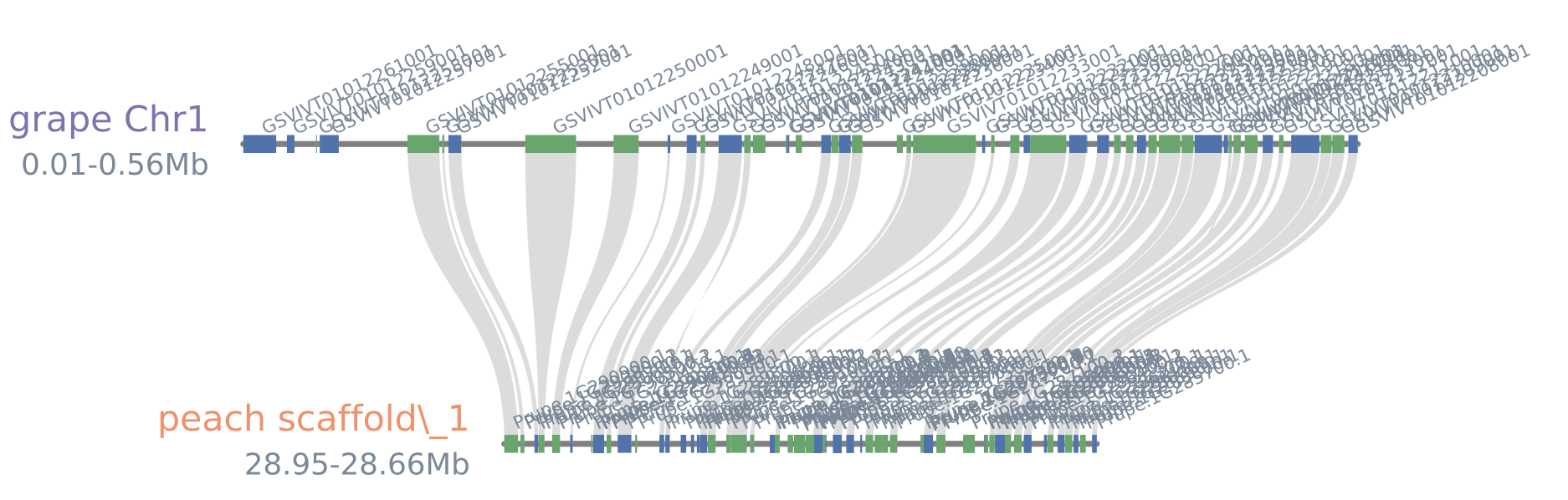
#blocks6.pdf(--genelabelsize=10 --genelabels=GSVIVT01012244001,Prupe.1G289800.1,GSVIVT01012225001,Prupe.1G288000.1)


