基于Pysam进行SAM/BAM文件数据统计
参考《Bioinfomatics Data Skill》第414页
一、首先安装pysam
$ pip install pysam
二、编写程序统计比对数据
以下代码具有一定参考价值,不如samtools stats直接好用。若是需要单独统计特殊信息,以下代码逻辑/框架具有一定参考价值。
import sys
import pysam
from collections import Counter
if len(sys.argv) < 2:
sys.exit("usage: alnstat.py in.bam")
fname = sys.argv[1]
bamfile = pysam.AlignmentFile(fname)
stats = Counter() for read in bamfile:
stats['total'] += 1
stats['qcfail'] += int(read.is_qcfail)
# record paired end info
stats['paired'] += int(read.is_paired) stats['read1'] += int(read.is_read1) stats['read2'] += int(read.is_read2)
if read.is_unmapped: stats['unmapped'] += 1
continue # other flags don't apply
# record if mapping quality <= 30
stats["mapping quality <= 30"] += int(read.mapping_quality <= 30) stats['mapped'] += 1
stats['proper pair'] += int(read.is_proper_pair)
# specify the output order, since dicts don't have order
output_order = ("total", "mapped", "unmapped", "paired", "read1", "read2", "proper pair", "qcfail",
"mapping quality <= 30")
# format output and print to standard out
for key in output_order:
format_args = (key, stats[key], 100*stats[key]/float(stats["total"]))
sys.stdout.write("%s: %d (%0.2f%%)\n" % format_args)
将上述代码存为alnstat.py,启动python对bam文件进行统计
$ python alnstat.py NA12891_CEU_sample.bam
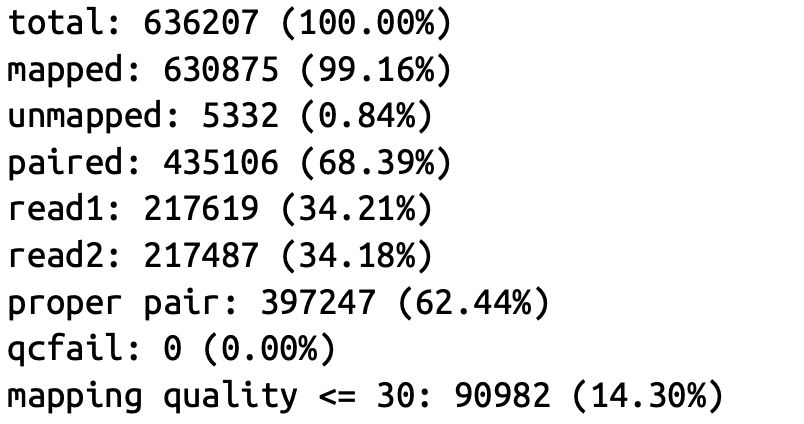
可得到如下结果: