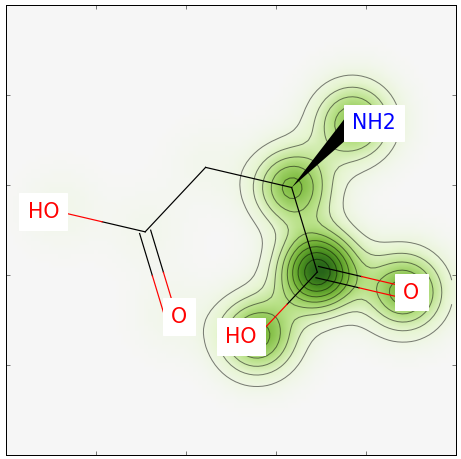
分子指纹相似度的可视化
rdkit有一个很炫酷的功能,那就是能可视化显示两个分子的相似性。
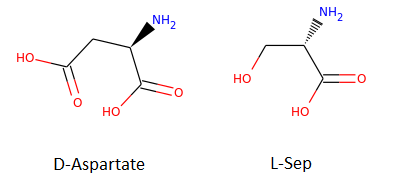
以下面两个分子为例:
计算相似度
from rdkit import Chem
from rdkit.Chem import AllChem, DataStructs
from rdkit.Chem.Fraggle import FraggleSim
# define TanimotoSim calculator for convinience.
def calctc(mol1,mol2):
fp1=AllChem.GetMorganFingerprintAsBitVect(mol1,2)
fp2=AllChem.GetMorganFingerprintAsBitVect(mol2,2)
return DataStructs.TanimotoSimilarity(fp1,fp2)
# make molecule from smiles.
mol=Chem.MolFromSmiles("N[C@H](CC(=O)O)C(=O)O")
mol2=Chem.MolFromSmiles("N[C@@H](CO)C(=O)O")
# calc. molecular similarity like ECFP4.
In [26]: calctc(mol,mol2)
Out[26]: 0.3333333333333333
#only N,C difference but low similarity !
# calc Fraggle sim.
In [27]:FraggleSim.GetFraggleSimilarity(mol,mol2)
Out[27]: (1.0, '[*]c1ccccc1.[*]c1ccccc1')
# near my feeling.
将相似度映射到分子图像
%matplotlib inline
%pylab inline
from IPython.display import Image
from rdkit.Chem import AllChem as Chem
from rdkit.Chem.Draw import IPythonConsole
from rdkit.Chem.Draw import SimilarityMaps
smiles1 = 'N[C@H](CC(=O)O)C(=O)O' #ZINC000000895218 (D-Aspartate)
smiles2 = 'N[C@@H](CO)C(=O)O' #ZINC000000895034 (L-Ser)
mol1 = Chem.MolFromSmiles(smiles1)
mol2 = Chem.MolFromSmiles(smiles2)
SimilarityMaps.GetSimilarityMapForFingerprint(mol2, mol1, SimilarityMaps.GetMorganFingerprint)
结果如下: