R语言中gene symbol 转换为ENTREZID, clusterprofile富集分析
001、
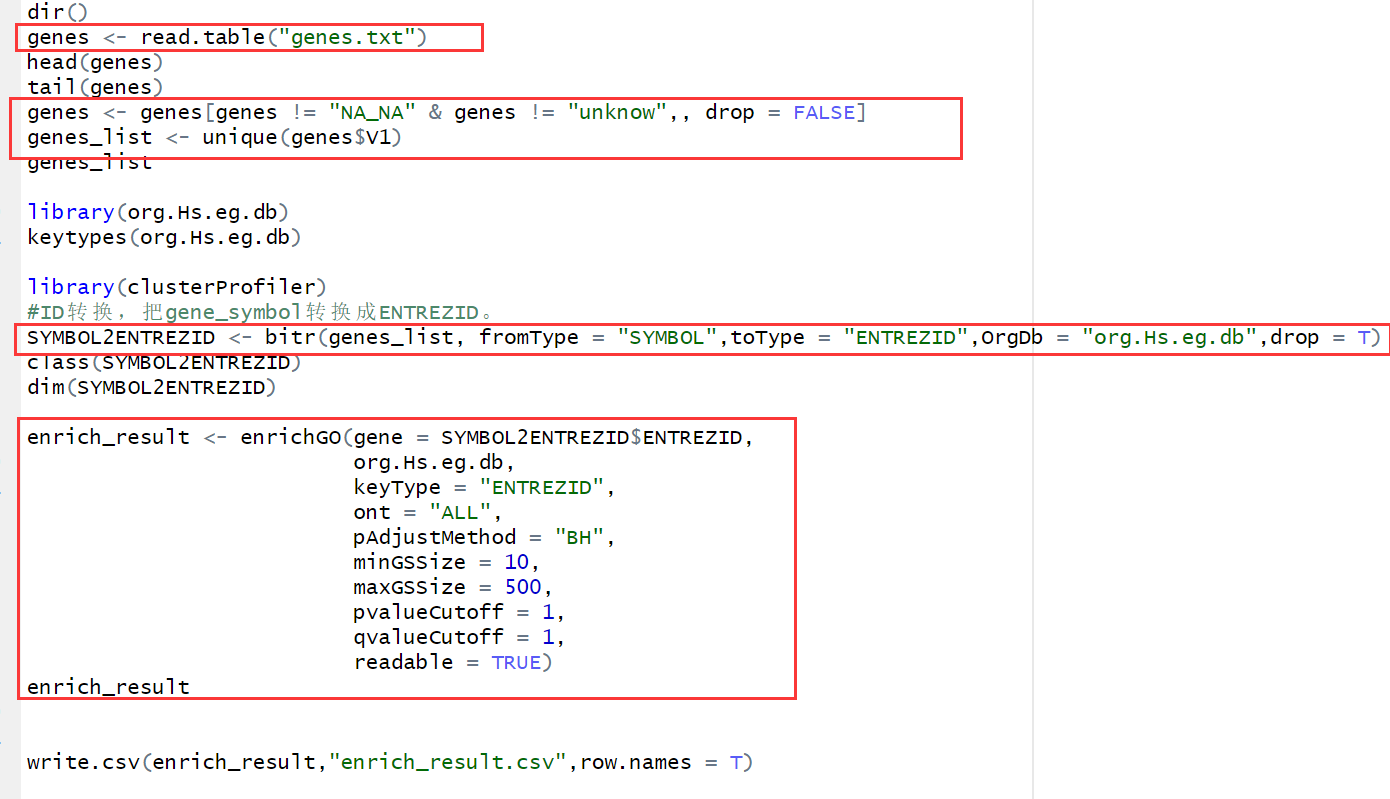
genes <- read.table("genes.txt") ## 读取基因symbol head(genes) tail(genes) genes <- genes[genes != "NA_NA" & genes != "unknow",, drop = FALSE] ## 去除无效信息(可选) genes_list <- unique(genes$V1) ## 去重 genes_list library(org.Hs.eg.db) keytypes(org.Hs.eg.db) library(clusterProfiler) #ID转换,把gene_symbol转换成ENTREZID。 SYMBOL2ENTREZID <- bitr(genes_list, fromType = "SYMBOL",toType = "ENTREZID",OrgDb = "org.Hs.eg.db",drop = T) ## 由symbol转换为ENTRZID class(SYMBOL2ENTREZID) dim(SYMBOL2ENTREZID) enrich_result <- enrichGO(gene = SYMBOL2ENTREZID$ENTREZID, ## 富集分析 org.Hs.eg.db, keyType = "ENTREZID", ont = "ALL", pAdjustMethod = "BH", minGSSize = 10, maxGSSize = 500, pvalueCutoff = 1, qvalueCutoff = 1, readable = TRUE) enrich_result write.csv(enrich_result,"enrich_result.csv",row.names = T) ## 保存富集分析结果
。






【推荐】国内首个AI IDE,深度理解中文开发场景,立即下载体验Trae
【推荐】编程新体验,更懂你的AI,立即体验豆包MarsCode编程助手
【推荐】抖音旗下AI助手豆包,你的智能百科全书,全免费不限次数
【推荐】轻量又高性能的 SSH 工具 IShell:AI 加持,快人一步
· 震惊!C++程序真的从main开始吗?99%的程序员都答错了
· 【硬核科普】Trae如何「偷看」你的代码?零基础破解AI编程运行原理
· 单元测试从入门到精通
· 上周热点回顾(3.3-3.9)
· winform 绘制太阳,地球,月球 运作规律
2023-10-04 linux 中设置用户间针对单一目录的权限
2023-10-04 python 中map函数
2023-10-04 samtools线程数对 sam文件转换为bam文件效率的影响
2023-10-04 bwa 线程数目对比对速度的影响
2023-10-04 python 中 re模块
2021-10-04 文本文件和二进制文件有什么区别?
2020-10-04 R语言中duplicated函数:去重复,取重复