Linux 中sed命令实现从gff文件中仅仅提取基因名称
001、
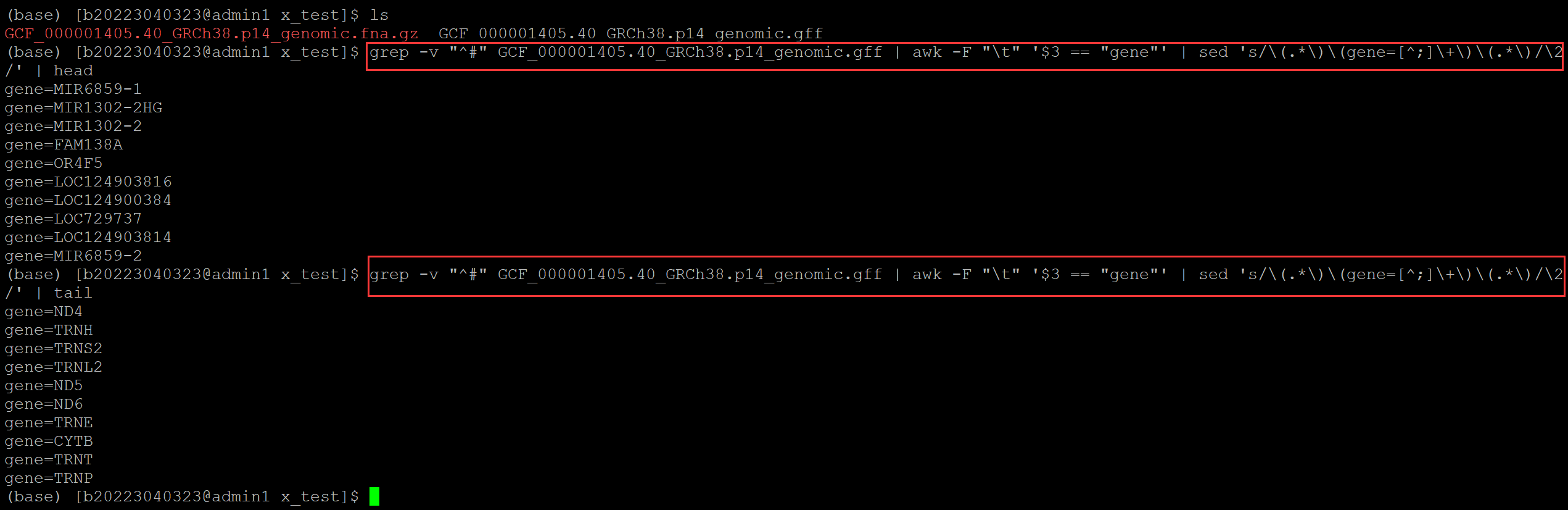
(base) [b20223040323@admin1 x_test]$ ls ## 测试gff文件 GCF_000001405.40_GRCh38.p14_genomic.fna.gz GCF_000001405.40_GRCh38.p14_genomic.gff (base) [b20223040323@admin1 x_test]$ grep -v "^#" GCF_000001405.40_GRCh38.p14_genomic.gff | awk -F "\t" '$3 == "gene"' | sed 's/\(.*\)\(gene=[^;]\+\)\(.*\)/\2/' | head ## 仅仅提取基因名称,并保留前10行 gene=MIR6859-1 gene=MIR1302-2HG gene=MIR1302-2 gene=FAM138A gene=OR4F5 gene=LOC124903816 gene=LOC124900384 gene=LOC729737 gene=LOC124903814 gene=MIR6859-2 (base) [b20223040323@admin1 x_test]$ grep -v "^#" GCF_000001405.40_GRCh38.p14_genomic.gff | awk -F "\t" '$3 == "gene"' | sed 's/\(.*\)\(gene=[^;]\+\)\(.*\)/\2/' | tail ## 仅仅提取基因名称,并保留后10行 gene=ND4 gene=TRNH gene=TRNS2 gene=TRNL2 gene=ND5 gene=ND6 gene=TRNE gene=CYTB gene=TRNT gene=TRNP
。
分类:
linux shell






【推荐】国内首个AI IDE,深度理解中文开发场景,立即下载体验Trae
【推荐】编程新体验,更懂你的AI,立即体验豆包MarsCode编程助手
【推荐】抖音旗下AI助手豆包,你的智能百科全书,全免费不限次数
【推荐】轻量又高性能的 SSH 工具 IShell:AI 加持,快人一步
· 震惊!C++程序真的从main开始吗?99%的程序员都答错了
· 【硬核科普】Trae如何「偷看」你的代码?零基础破解AI编程运行原理
· 单元测试从入门到精通
· 上周热点回顾(3.3-3.9)
· winform 绘制太阳,地球,月球 运作规律
2022-05-03 R语言中seq函数的用法
2022-05-03 R语言中fread函数中colClasses = "character"选项
2022-05-03 intel CPU、AMD CPU
2021-05-03 python中创建字典、字典的访问
2021-05-03 TypeError: 'dict' object is not callable
2021-05-03 c语言 5-9
2021-05-03 c语言 5-12