linux 中awk命令实现根据一列数值的大小筛选指定列多个类别的最大或者最小项
001、
(base) [b20223040323@admin1 test2]$ cat 003.txt ## 测试数据如下,第一列有多个项,且部分项有重复,实现根据第三列筛选出最大的项 ID=gene-TRNAC-GCA rna-TRNAC-GCA 72 ID=gene-ATP5O rna-XM_005674665.3 793 ID=gene-ITSN1 rna-XR_001917533.1 4437 ID=gene-ITSN1 rna-XM_018043229.1 9110 ID=gene-ITSN1 rna-XM_018041619.1 9323 ID=gene-ITSN1 rna-XM_018042670.1 9247 ID=gene-ITSN1 rna-XM_018042157.1 9141 ID=gene-ITSN1 rna-XM_018043907.1 5462 ID=gene-CRYZL1 rna-XM_005674667.3 1609 ID=gene-CRYZL1 rna-XM_013962888.2 1588 ID=gene-CRYZL1 rna-XM_005674668.3 1600 ID=gene-CRYZL1 rna-XM_013962952.2 1749 ID=gene-CRYZL1 rna-XM_005674666.3 1745 ID=gene-DONSON rna-XM_018045976.1 2568 ID=gene-SON rna-XR_001295570.2 8568 ID=gene-SON rna-XM_005674676.3 8488 ID=gene-SON rna-XM_005674677.3 8409 ID=gene-SON rna-XM_005674679.3 7645 ID=gene-SON rna-XM_005674680.3 8005 ID=gene-GART rna-XM_005674671.3 4934
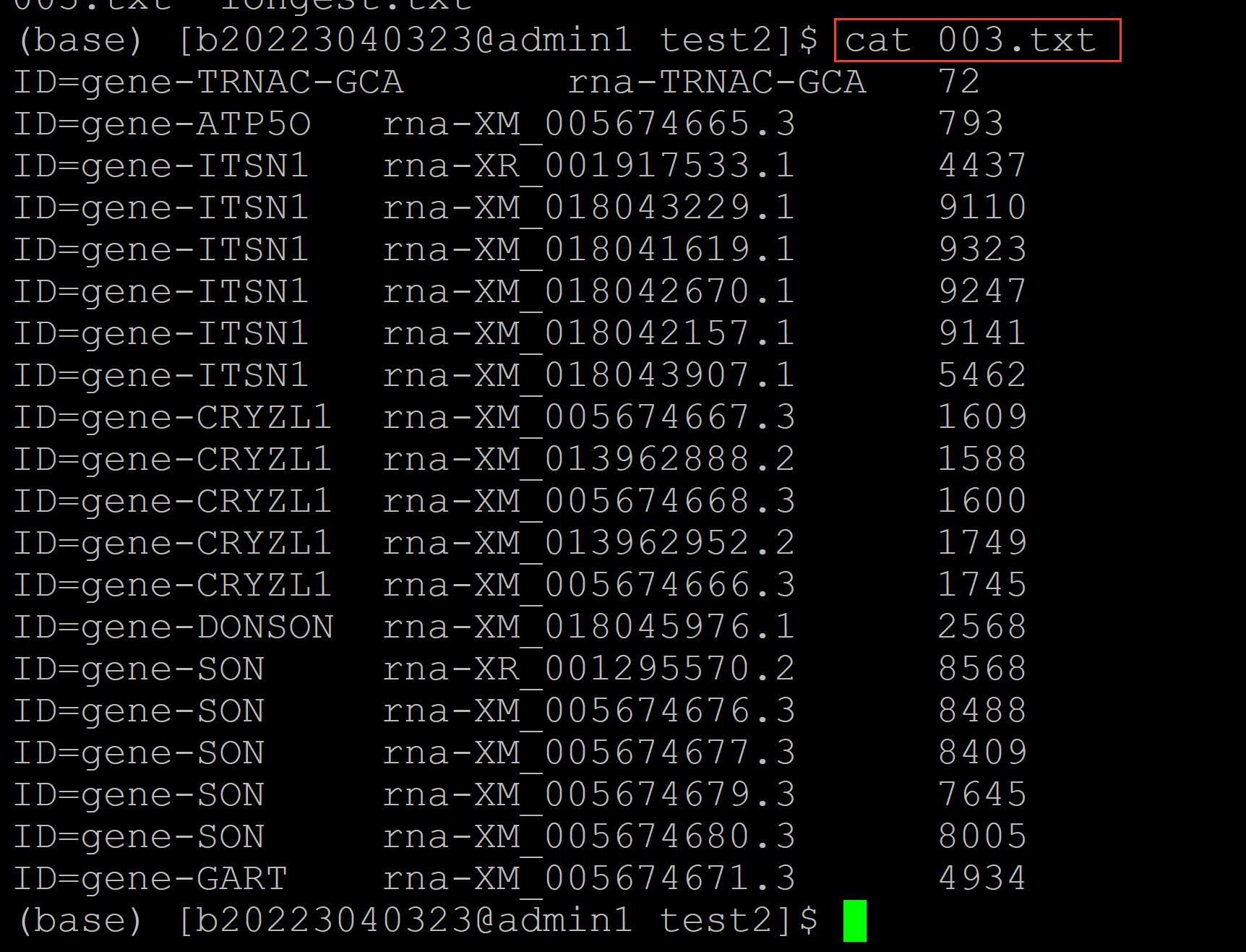
002、输出最大项的实现方法如下:
(base) [b20223040323@admin1 test2]$ awk '{if(ay1[$1] == "") {ay1[$1] = $3; ay2[$1] = $0};if(ay1[$1] < $3){ay1[$1] = $3; ay2[$1] = $0} } END{for(i in ay1) {print ay2[i]}}' 003.txt ID=gene-GART rna-XM_005674671.3 4934 ## 借助awk 数组来实现; 基本思想利用awk循环对第三列数值进行判断;出现最大值,则将当前行存储在ay2数组中 ID=gene-TRNAC-GCA rna-TRNAC-GCA 72 ID=gene-SON rna-XR_001295570.2 8568 ID=gene-DONSON rna-XM_018045976.1 2568 ID=gene-CRYZL1 rna-XM_013962952.2 1749 ID=gene-ATP5O rna-XM_005674665.3 793 ID=gene-ITSN1 rna-XM_018041619.1 9323
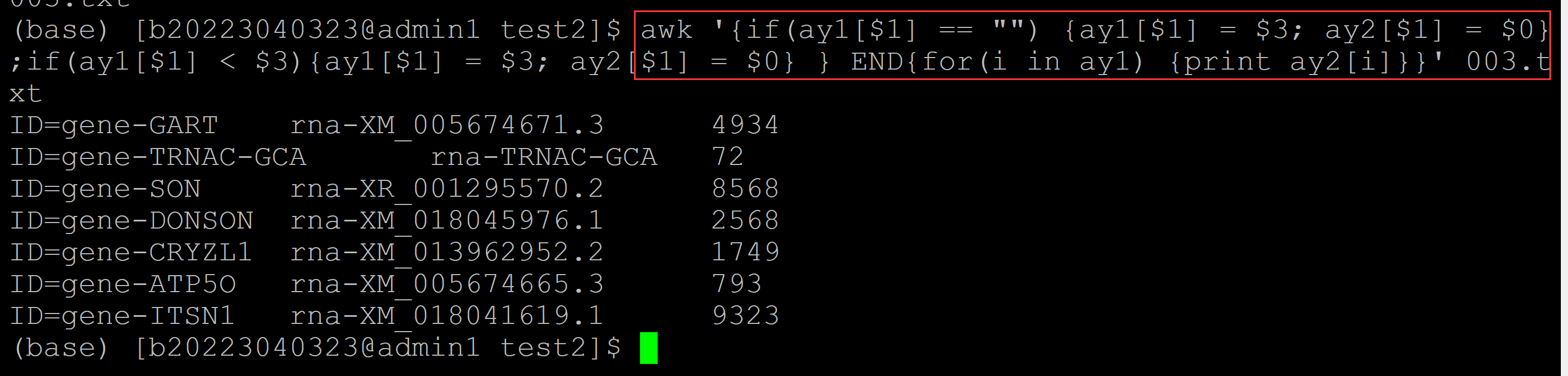
003、输出最小项的实现方法如下:
(base) [b20223040323@admin1 test2]$ awk '{if(ay1[$1] == "") {ay1[$1] = $3; ay2[$1] = $0}; if(ay1[$1] > $3){ay1[$1] = $3; ay2[$1] = $0}} END {for(i in ay1) {print ay2[i]}}' 003.txt ID=gene-GART rna-XM_005674671.3 4934 ## 实现的方法与输出最大项方法一致 ID=gene-TRNAC-GCA rna-TRNAC-GCA 72 ID=gene-SON rna-XM_005674679.3 7645 ID=gene-DONSON rna-XM_018045976.1 2568 ID=gene-CRYZL1 rna-XM_013962888.2 1588 ID=gene-ATP5O rna-XM_005674665.3 793 ID=gene-ITSN1 rna-XR_001917533.1 4437
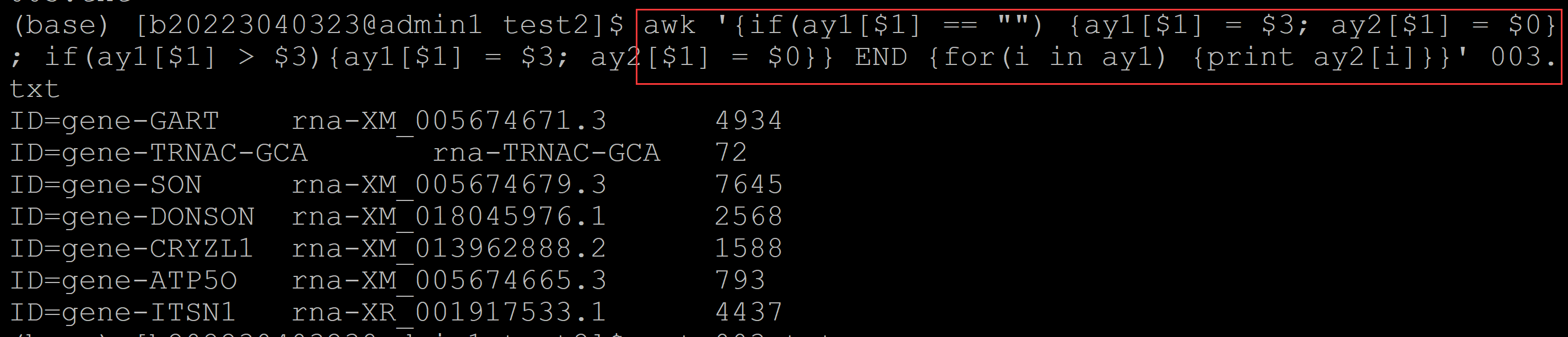
004、如何保持第一列的顺序
a、输出最大项,并保持第一列的顺序
(base) [b20223040323@admin1 test2]$ awk '{if(ay1[$1] == "") {tmp++; ay1[$1] = $3; ay2[tmp] = $0};if(ay1[$1] < $3){ay1[$1] = $3; ay2[tmp] = $0} } END{for(i = 1; i <= length(ay2); i++) {print ay2[i]}}' 003.txt ID=gene-TRNAC-GCA rna-TRNAC-GCA 72 ## 利用tmp计数; 最后按照for数值循环输出 ID=gene-ATP5O rna-XM_005674665.3 793 ID=gene-ITSN1 rna-XM_018041619.1 9323 ID=gene-CRYZL1 rna-XM_013962952.2 1749 ID=gene-DONSON rna-XM_018045976.1 2568 ID=gene-SON rna-XR_001295570.2 8568 ID=gene-GART rna-XM_005674671.3 4934
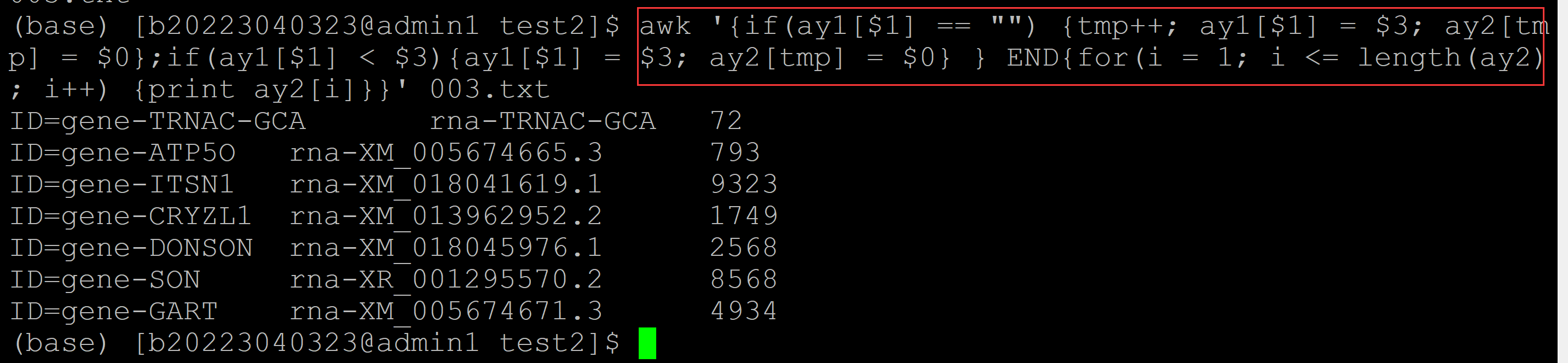
b、输出最小项,并保持第一列的顺序
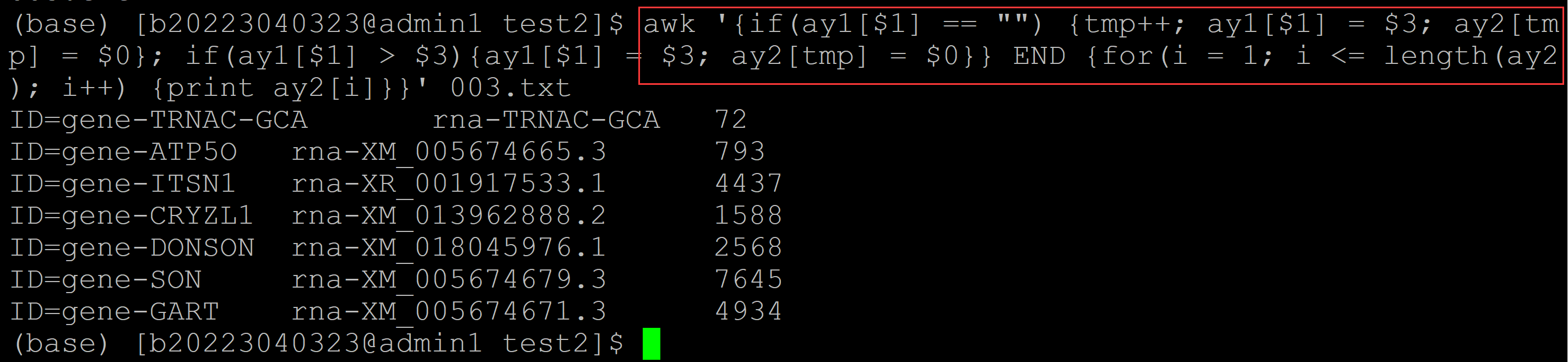
(base) [b20223040323@admin1 test2]$ awk '{if(ay1[$1] == "") {tmp++; ay1[$1] = $3; ay2[tmp] = $0}; if(ay1[$1] > $3){ay1[$1] = $3; ay2[tmp] = $0}} END {for(i = 1; i <= length(ay2); i++) {print ay2[i]}}' 003.txt ID=gene-TRNAC-GCA rna-TRNAC-GCA 72 ## 输出最小项,并保持第一列的顺序 ID=gene-ATP5O rna-XM_005674665.3 793 ID=gene-ITSN1 rna-XR_001917533.1 4437 ID=gene-CRYZL1 rna-XM_013962888.2 1588 ID=gene-DONSON rna-XM_018045976.1 2568 ID=gene-SON rna-XM_005674679.3 7645 ID=gene-GART rna-XM_005674671.3 4934
。
分类:
linux shell






【推荐】国内首个AI IDE,深度理解中文开发场景,立即下载体验Trae
【推荐】编程新体验,更懂你的AI,立即体验豆包MarsCode编程助手
【推荐】抖音旗下AI助手豆包,你的智能百科全书,全免费不限次数
【推荐】轻量又高性能的 SSH 工具 IShell:AI 加持,快人一步
· 震惊!C++程序真的从main开始吗?99%的程序员都答错了
· 【硬核科普】Trae如何「偷看」你的代码?零基础破解AI编程运行原理
· 单元测试从入门到精通
· 上周热点回顾(3.3-3.9)
· winform 绘制太阳,地球,月球 运作规律