python中计算dna序列的GC含量
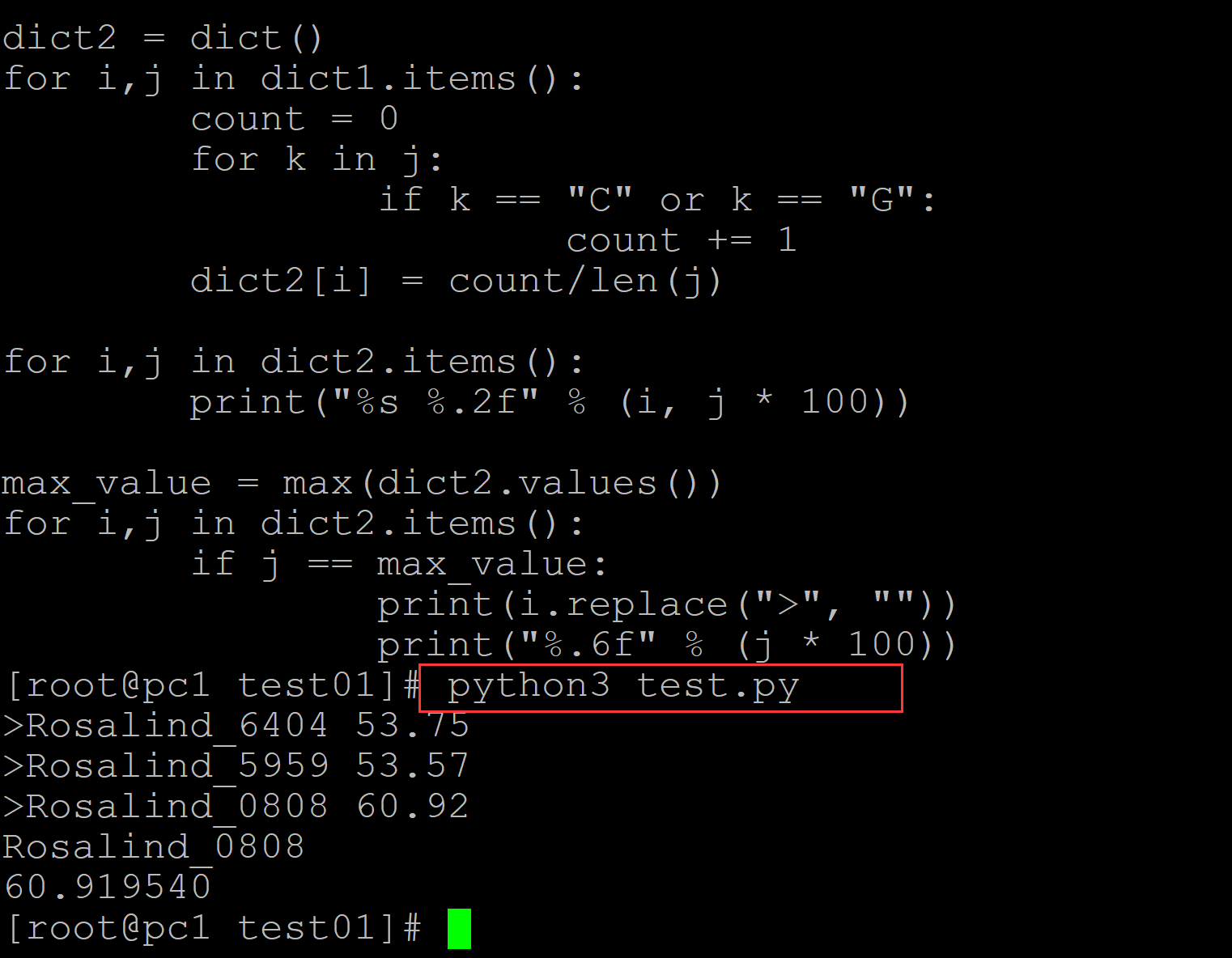
001、对G、C计数进行统计
[root@pc1 test01]# ls a.fa test.py [root@pc1 test01]# cat a.fa ## 测试DNA序列 >Rosalind_6404 CCTGCGGAAGATCGGCACTAGAATAGCCAGAACCGTTTCTCTGAGGCTTCCGGCCTTCCC TCCCACTAATAATTCTGAGG >Rosalind_5959 CCATCGGTAGCGCATCCTTAGTCCAATTAAGTCCCTATCCAGGCGCTCCGCCGAAGGTCT ATATCCATTTGTCAGCAGACACGC >Rosalind_0808 CCACCCTCGTGGTATGGCTAGGCATTCAGGAACCGGAGAACGCTTCAGACCAGCCCGGAC TGGGAACCTGCGGGCAGTAGGTGGAAT [root@pc1 test01]# cat test.py ## 统计程序 #!/usr/bin/env python # -*- coding: utf-8 -*- in_file = open("a.fa", "r") dict1 = dict() for i in in_file: i = i.strip() if i.startswith(">"): temp = i dict1[temp] = str() else: dict1[temp] += i in_file.close() dict2 = dict() for i,j in dict1.items(): count = 0 for k in j: if k == "C" or k == "G": count += 1 dict2[i] = count/len(j) for i,j in dict2.items(): print("%s %.2f" % (i, j * 100)) max_value = max(dict2.values()) for i,j in dict2.items(): if j == max_value: print(i.replace(">", "")) print("%.6f" % (j * 100)) [root@pc1 test01]# python3 test.py ## 输出结果 >Rosalind_6404 53.75 >Rosalind_5959 53.57 >Rosalind_0808 60.92 Rosalind_0808 60.919540
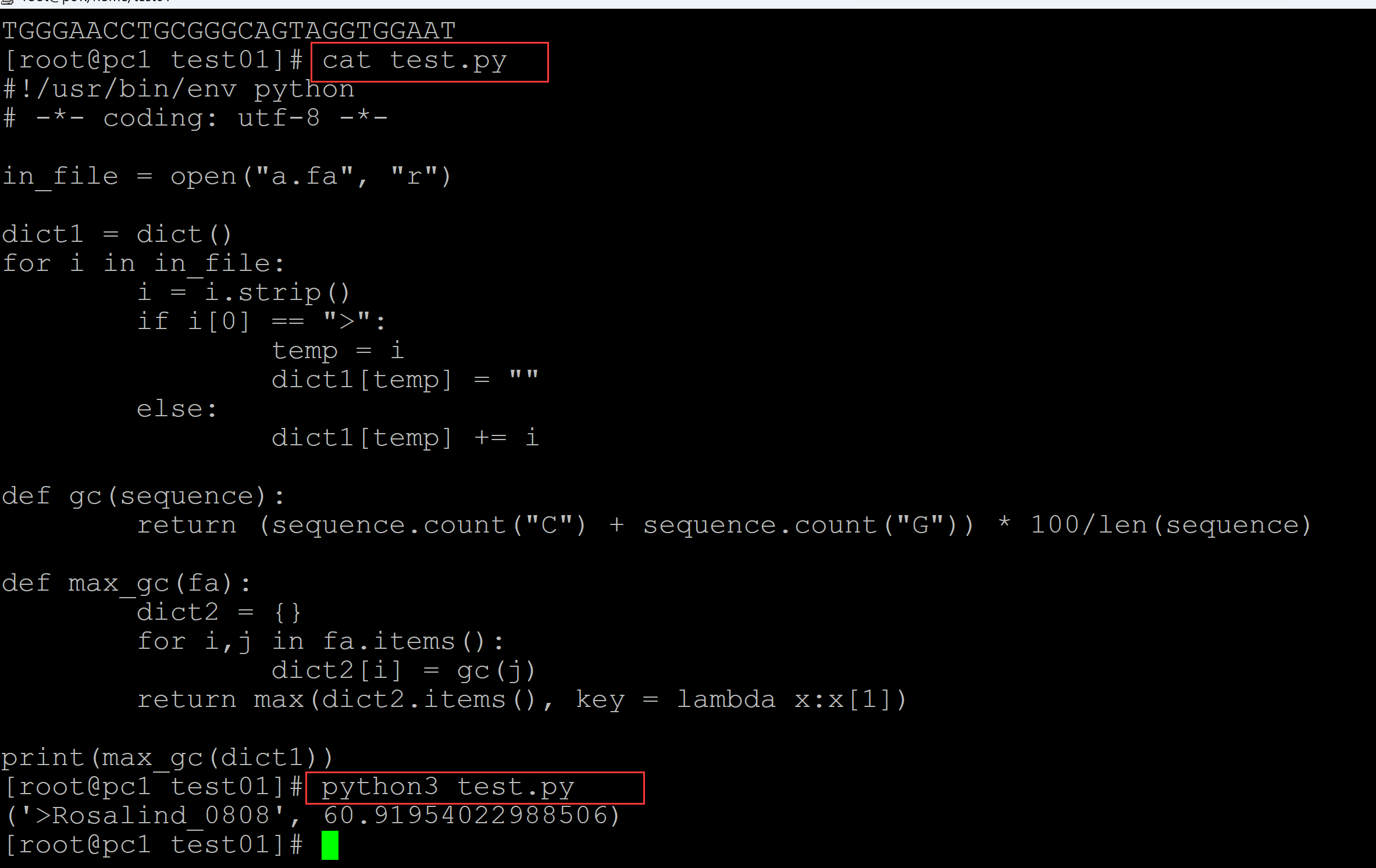
002、利用函数结构实现
[root@pc1 test01]# ls a.fa test.py [root@pc1 test01]# cat a.fa ## 测试fasta文件 >Rosalind_6404 CCTGCGGAAGATCGGCACTAGAATAGCCAGAACCGTTTCTCTGAGGCTTCCGGCCTTCCC TCCCACTAATAATTCTGAGG >Rosalind_5959 CCATCGGTAGCGCATCCTTAGTCCAATTAAGTCCCTATCCAGGCGCTCCGCCGAAGGTCT ATATCCATTTGTCAGCAGACACGC >Rosalind_0808 CCACCCTCGTGGTATGGCTAGGCATTCAGGAACCGGAGAACGCTTCAGACCAGCCCGGAC TGGGAACCTGCGGGCAGTAGGTGGAAT [root@pc1 test01]# cat test.py ## 统计程序 #!/usr/bin/env python # -*- coding: utf-8 -*- in_file = open("a.fa", "r") dict1 = dict() for i in in_file: i = i.strip() if i[0] == ">": temp = i dict1[temp] = "" else: dict1[temp] += i def gc(sequence): return (sequence.count("C") + sequence.count("G")) * 100/len(sequence) def max_gc(fa): dict2 = {} for i,j in fa.items(): dict2[i] = gc(j) return max(dict2.items(), key = lambda x:x[1]) print(max_gc(dict1)) [root@pc1 test01]# python3 test.py ## 统计结果 ('>Rosalind_0808', 60.91954022988506)
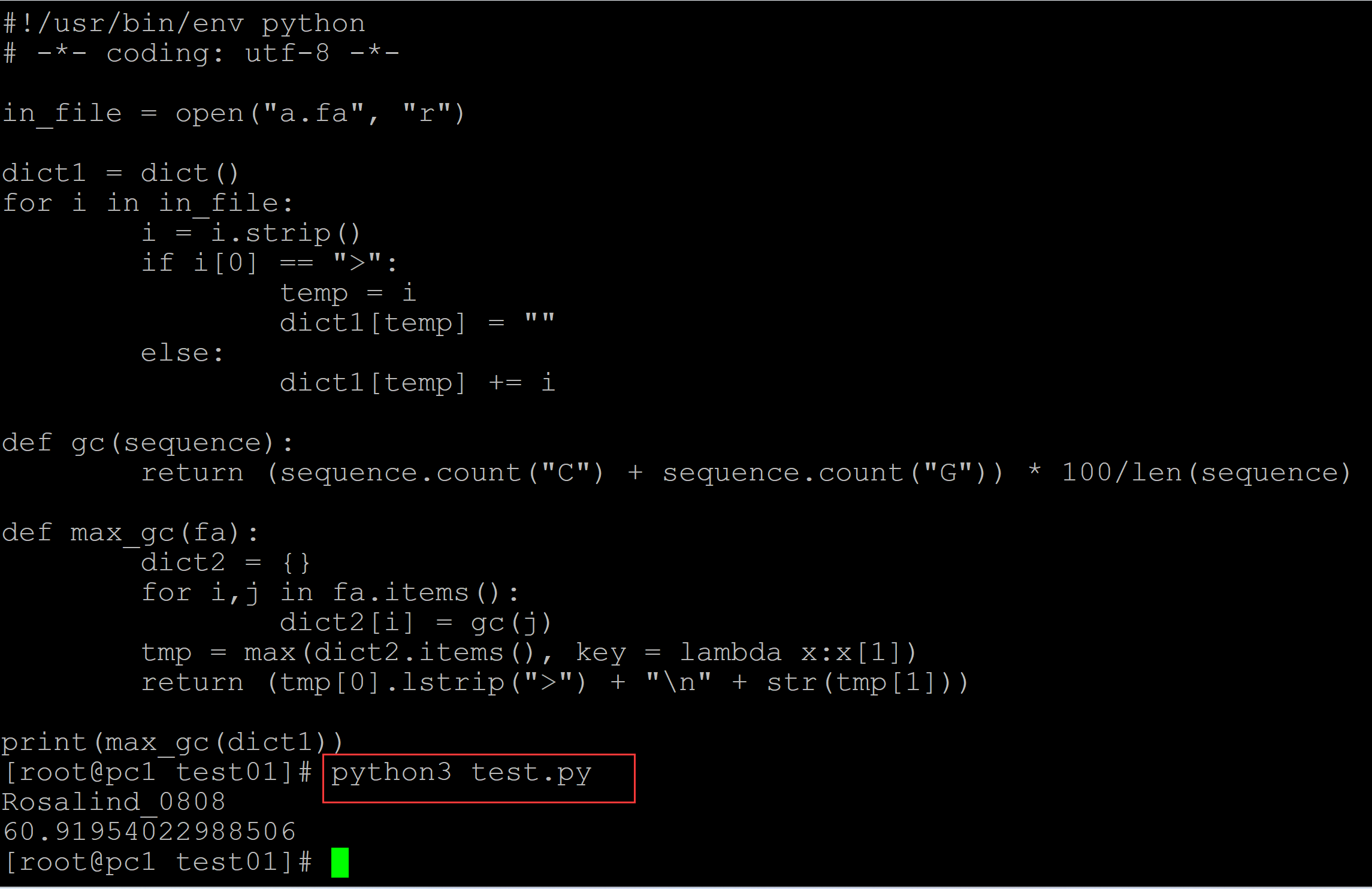
003、改进
[root@pc1 test01]# ls a.fa test.py [root@pc1 test01]# cat a.fa ## 测试DNA序列 >Rosalind_6404 CCTGCGGAAGATCGGCACTAGAATAGCCAGAACCGTTTCTCTGAGGCTTCCGGCCTTCCC TCCCACTAATAATTCTGAGG >Rosalind_5959 CCATCGGTAGCGCATCCTTAGTCCAATTAAGTCCCTATCCAGGCGCTCCGCCGAAGGTCT ATATCCATTTGTCAGCAGACACGC >Rosalind_0808 CCACCCTCGTGGTATGGCTAGGCATTCAGGAACCGGAGAACGCTTCAGACCAGCCCGGAC TGGGAACCTGCGGGCAGTAGGTGGAAT [root@pc1 test01]# cat test.py ## 计算程序 #!/usr/bin/env python # -*- coding: utf-8 -*- in_file = open("a.fa", "r") dict1 = dict() for i in in_file: i = i.strip() if i[0] == ">": temp = i dict1[temp] = "" else: dict1[temp] += i def gc(sequence): return (sequence.count("C") + sequence.count("G")) * 100/len(sequence) def max_gc(fa): dict2 = {} for i,j in fa.items(): dict2[i] = gc(j) tmp = max(dict2.items(), key = lambda x:x[1]) return (tmp[0].lstrip(">") + "\n" + str(tmp[1])) print(max_gc(dict1)) [root@pc1 test01]# python3 test.py ## 计算结果 Rosalind_0808 60.91954022988506
。






【推荐】国内首个AI IDE,深度理解中文开发场景,立即下载体验Trae
【推荐】编程新体验,更懂你的AI,立即体验豆包MarsCode编程助手
【推荐】抖音旗下AI助手豆包,你的智能百科全书,全免费不限次数
【推荐】轻量又高性能的 SSH 工具 IShell:AI 加持,快人一步
· 震惊!C++程序真的从main开始吗?99%的程序员都答错了
· 【硬核科普】Trae如何「偷看」你的代码?零基础破解AI编程运行原理
· 单元测试从入门到精通
· 上周热点回顾(3.3-3.9)
· winform 绘制太阳,地球,月球 运作规律
2022-08-28 R语言中快捷键 选中光标的文件头的所有内容、及选种光标到文件尾的所有内容
2022-08-28 R语言中快捷键 选中光标所在行
2022-08-28 R语言中删除光标到行首、光标到行末的快捷键
2022-08-28 seurat 单细胞数据分析中MinMax函数
2022-08-28 R语言中依据数据框元素筛选元素
2021-08-28 c primer plus 8编程练习