linux 中shell 脚本将 gff文件转换为bed文件
001、
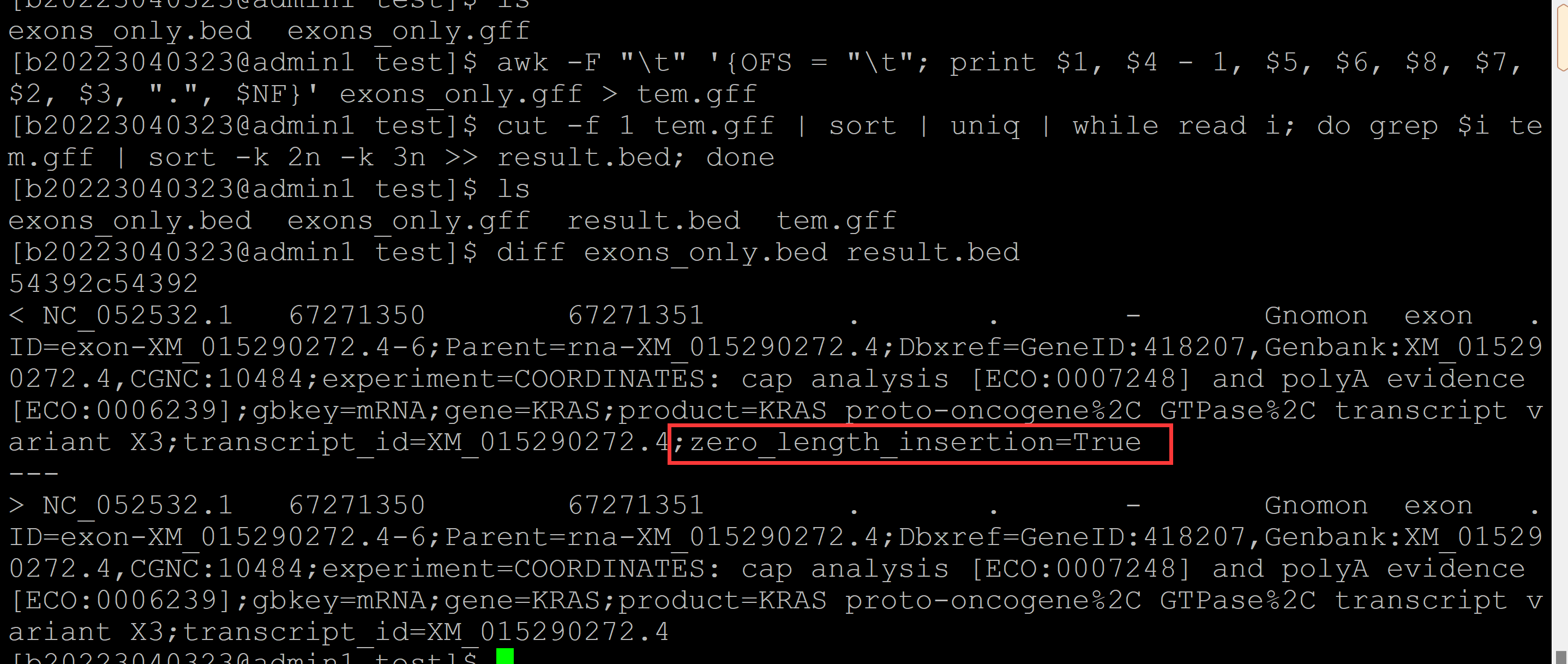
[b20223040323@admin1 test]$ ls ## 测试gff文件 exons_only.gff [b20223040323@admin1 test]$ gff2bed <exons_only.gff > exons_only.bed ## gff2bed模块转换 Warning: If your Wiggle data is a significant portion of available system memory, use the --max-mem and --sort-tmpdir options, or use --do-not-sort to disable post-conversion sorting. See --help for more information. [b20223040323@admin1 test]$ ls ## 转换结果 exons_only.bed exons_only.gff [b20223040323@admin1 test]$ awk -F "\t" '{OFS = "\t"; print $1, $4 - 1, $5, $6, $8, $7, $2, $3, ".", $NF}' exons_only.gff > tem.gff ## 列的重排, [b20223040323@admin1 test]$ cut -f 1 tem.gff | sort | uniq | while read i; do grep $i tem.gff | sort -k 2n -k 3n >> result.bed; done ## 排序 [b20223040323@admin1 test]$ ls ## 结果文件 exons_only.bed exons_only.gff result.bed tem.gff [b20223040323@admin1 test]$ diff exons_only.bed result.bed ## 比较gff2bed模块和shell脚本的结果, 有一行差异?? 54392c54392 < NC_052532.1 67271350 67271351 . . - Gnomon exon .ID=exon-XM_015290272.4-6;Parent=rna-XM_015290272.4;Dbxref=GeneID:418207,Genbank:XM_015290272.4,CGNC:10484;experiment=COORDINATES: cap analysis [ECO:0007248] and polyA evidence [ECO:0006239];gbkey=mRNA;gene=KRAS;product=KRAS proto-oncogene%2C GTPase%2C transcript variant X3;transcript_id=XM_015290272.4;zero_length_insertion=True --- > NC_052532.1 67271350 67271351 . . - Gnomon exon .ID=exon-XM_015290272.4-6;Parent=rna-XM_015290272.4;Dbxref=GeneID:418207,Genbank:XM_015290272.4,CGNC:10484;experiment=COORDINATES: cap analysis [ECO:0007248] and polyA evidence [ECO:0006239];gbkey=mRNA;gene=KRAS;product=KRAS proto-oncogene%2C GTPase%2C transcript variant X3;transcript_id=XM_015290272.4
分类:
生信






【推荐】国内首个AI IDE,深度理解中文开发场景,立即下载体验Trae
【推荐】编程新体验,更懂你的AI,立即体验豆包MarsCode编程助手
【推荐】抖音旗下AI助手豆包,你的智能百科全书,全免费不限次数
【推荐】轻量又高性能的 SSH 工具 IShell:AI 加持,快人一步
· 震惊!C++程序真的从main开始吗?99%的程序员都答错了
· 【硬核科普】Trae如何「偷看」你的代码?零基础破解AI编程运行原理
· 单元测试从入门到精通
· 上周热点回顾(3.3-3.9)
· winform 绘制太阳,地球,月球 运作规律
2020-11-12 ansible