seurat 单细胞数据分析中 VizDimLoadings 函数
前期处理:https://www.jianshu.com/p/fef17a1babc2
#可视化对每个主成分影响比较大的基因集
001、
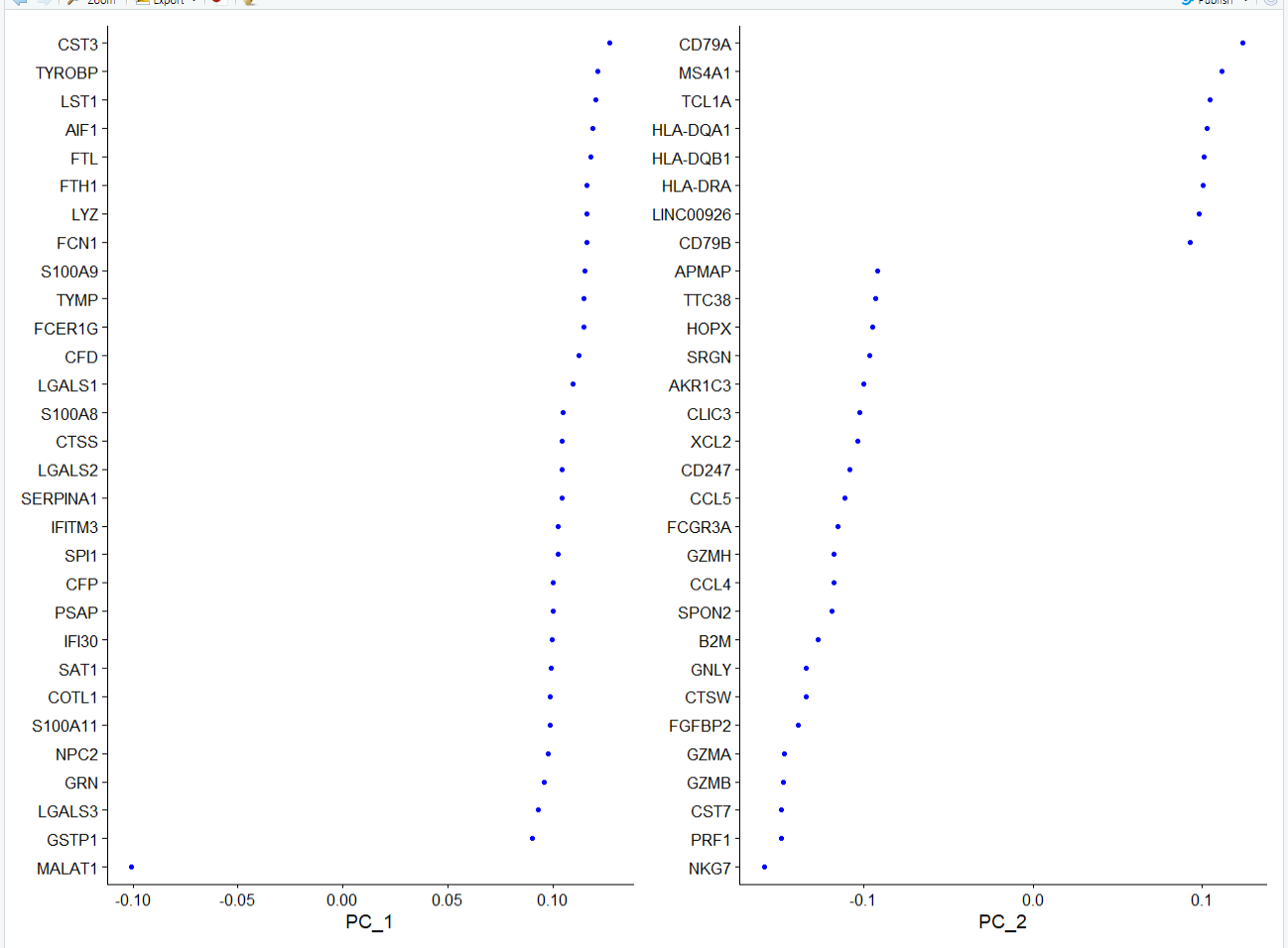
dat <- pbmc[["pca"]]@feature.loadings ## 绘图数据 dat[1:3, 1:3] par(mai = c(1, 1, 1, 1),mgp = c(2.5,0.7,0)) par(mfrow = c(1,2)) datx <- data.frame(PC_1 = dat[,1]) datx <- datx[order(abs(datx$PC_1), decreasing = T),,drop = F] %>% head(30) datx <- datx[order(datx$PC_1, decreasing = T),, drop = F] datx$num <- nrow(datx):1 plot(datx$PC_1, datx$num, yaxt = "n", cex = 2,pch = 19, ylab = "", xlab = "PC_1", col = "purple") axis(2, at = 1:30, labels = rev(rownames(datx)), las = 2) datx <- data.frame(PC_2 = dat[,2]) datx <- datx[order(abs(datx$PC_2), decreasing = T),,drop = F] %>% head(30) datx <- datx[order(datx$PC_2, decreasing = T),, drop = F] datx$num <- nrow(datx):1 plot(datx$PC_2, datx$num, yaxt = "n", cex = 2, pch = 19, ylab = "", xlab = "PC_2", col = "purple") axis(2, at = 1:30, labels = rev(rownames(datx)), las = 2)

标准结果:
VizDimLoadings(pbmc, dims = 1:2, reduction = "pca")



 浙公网安备 33010602011771号
浙公网安备 33010602011771号