Seurat4.0单细胞数据分析 细胞pca值的计算验证
001、前期处理:
https://www.jianshu.com/p/4f7aeae81ef1

基本过程如上图:
001、左图数据为scale.data, pbmc[["RNA"]]@scale.data
002、中图数据为feature.loadings, pbmc[["pca"]]@feature.loadings
003、右图数据为:t(pbmc[["pca"]]@cell.embeddings)
002、验证
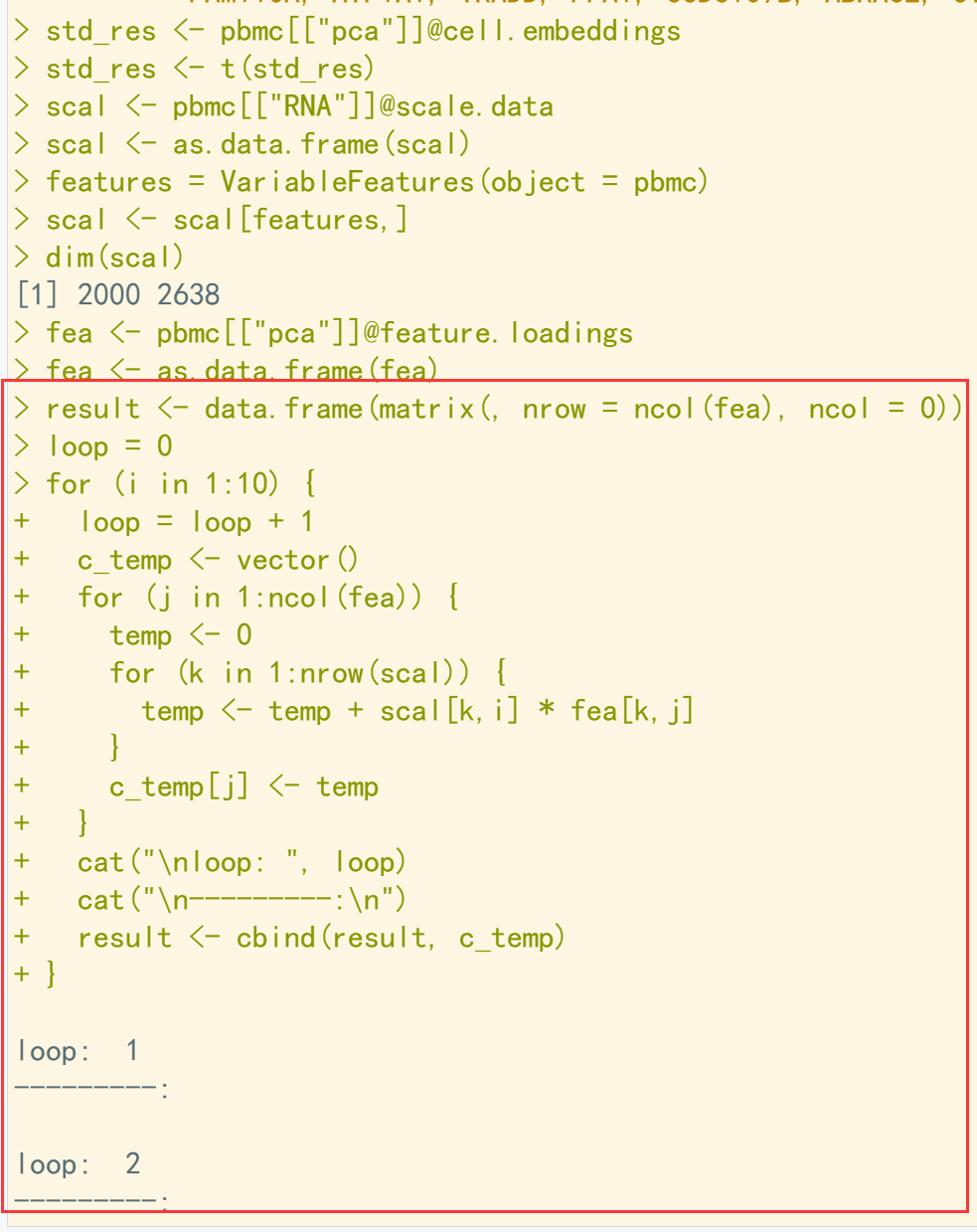
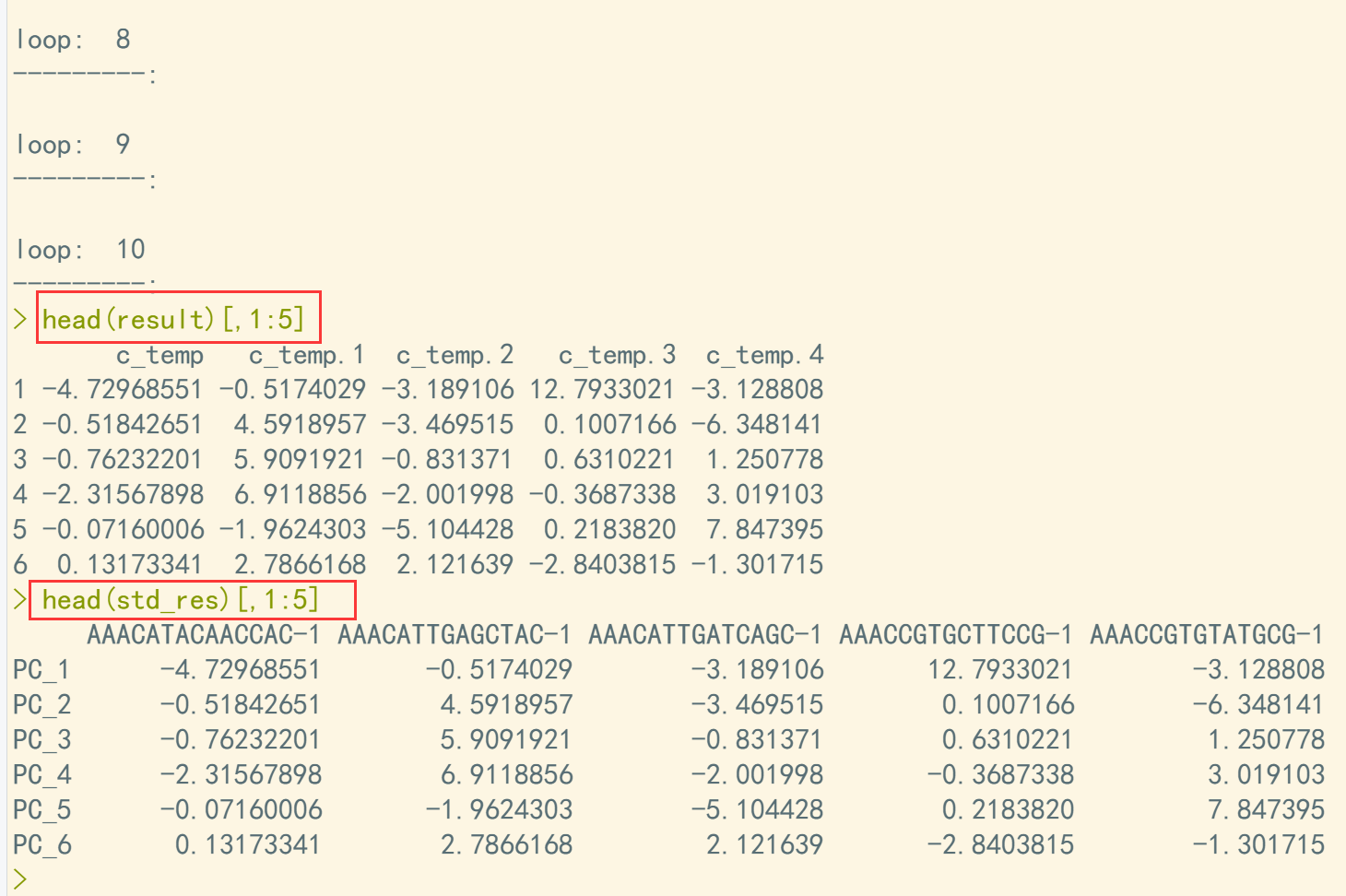
pbmc <- RunPCA(pbmc, features = VariableFeatures(object = pbmc)) ## 运行PCA std_res <- pbmc[["pca"]]@cell.embeddings std_res <- t(std_res) ## 将细胞pca结果转置 scal <- pbmc[["RNA"]]@scale.data scal <- as.data.frame(scal) features = VariableFeatures(object = pbmc) scal <- scal[features,] ## 提取scale数据) dim(scal) fea <- pbmc[["pca"]]@feature.loadings fea <- as.data.frame(fea) ## 提取feature数据 result <- data.frame(matrix(, nrow = ncol(fea), ncol = 0)) loop = 0 for (i in 1:10) { ## 利用循环结果计算PCA值,这里计算10个循环 loop = loop + 1 c_temp <- vector() for (j in 1:ncol(fea)) { temp <- 0 for (k in 1:nrow(scal)) { temp <- temp + scal[k,i] * fea[k,j] } c_temp[j] <- temp } cat("\nloop: ", loop) cat("\n---------:\n") result <- cbind(result, c_temp) } head(result)[,1:5] ## 对比计算结果和标准计算结果 head(std_res)[,1:5]
二、利用矩阵乘法实现
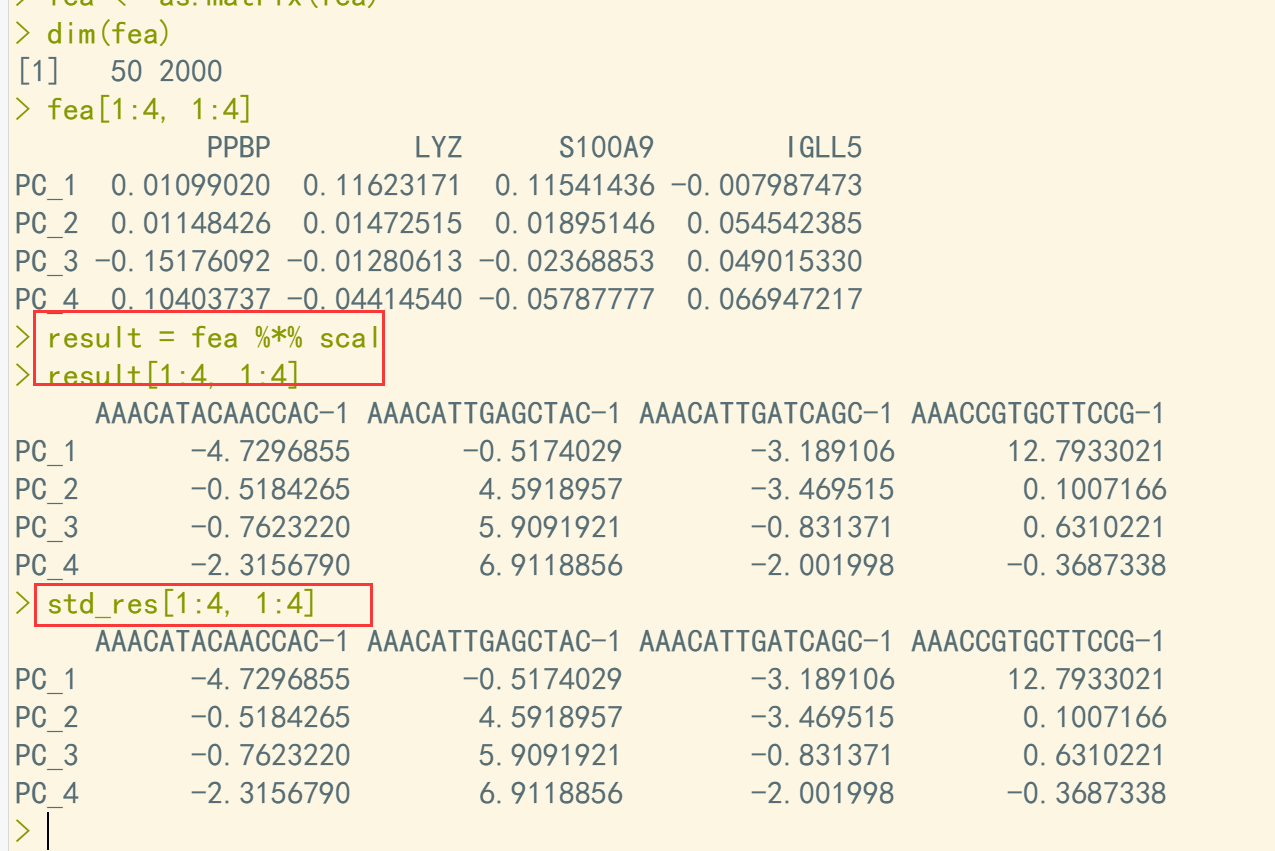
pbmc <- RunPCA(pbmc, features = VariableFeatures(object = pbmc)) std_res <- pbmc[["pca"]]@cell.embeddings std_res <- t(std_res) ## 标准答案 scal <- pbmc[["RNA"]]@scale.data scal <- as.data.frame(scal) features = VariableFeatures(object = pbmc) scal <- scal[features,] scal = as.matrix(scal) scal[1:4, 1:4] fea <- pbmc[["pca"]]@feature.loadings fea <- as.data.frame(fea) fea <- t(fea) fea <- as.matrix(fea) dim(fea) fea[1:4, 1:4] result = fea %*% scal ## 利用矩阵相乘计算PCA result[1:4, 1:4] ## 结果比对 std_res[1:4, 1:4]
参考:https://www.bilibili.com/video/BV1Qf4y1s7h1?p=2&vd_source=9f2c87b6a7fd328f19dccf7b7b1f8c1a






【推荐】国内首个AI IDE,深度理解中文开发场景,立即下载体验Trae
【推荐】编程新体验,更懂你的AI,立即体验豆包MarsCode编程助手
【推荐】抖音旗下AI助手豆包,你的智能百科全书,全免费不限次数
【推荐】轻量又高性能的 SSH 工具 IShell:AI 加持,快人一步
· 震惊!C++程序真的从main开始吗?99%的程序员都答错了
· 【硬核科普】Trae如何「偷看」你的代码?零基础破解AI编程运行原理
· 单元测试从入门到精通
· 上周热点回顾(3.3-3.9)
· winform 绘制太阳,地球,月球 运作规律