GWAS logistic + 协变量 回归分析
001、plink
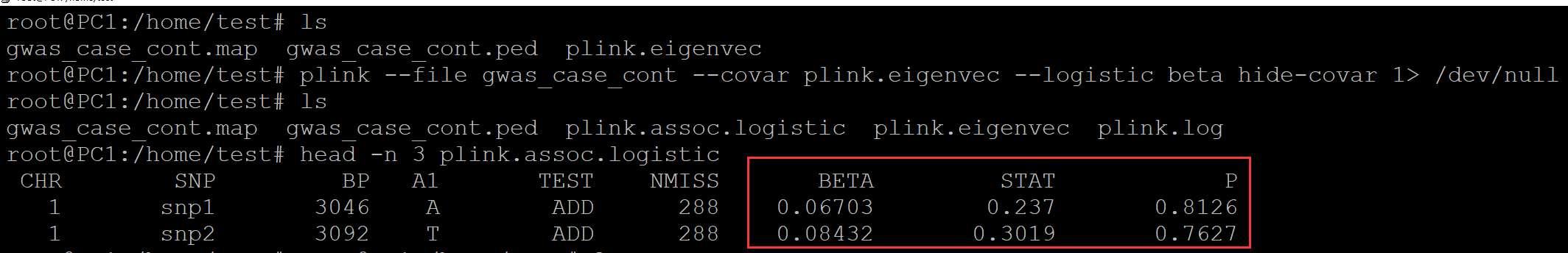
root@PC1:/home/test# ls gwas_case_cont.map gwas_case_cont.ped root@PC1:/home/test# plink --file gwas_case_cont --pca 3 1> /dev/null ## 生成PCA协变量 root@PC1:/home/test# ls gwas_case_cont.map gwas_case_cont.ped plink.eigenval plink.eigenvec plink.log root@PC1:/home/test# head -n 3 plink.eigenvec | column -t A1 A1 0.0161698 0.108569 -0.0182556 A2 A2 -0.0582408 0.00512212 0.0141237 A3 A3 -0.0363562 -0.0640895 0.0172107 root@PC1:/home/test# plink --file gwas_case_cont --covar plink.eigenvec --logistic hide-covar 1> /dev.null ## 逻辑回归, 添加协变量 root@PC1:/home/test# ls gwas_case_cont.map gwas_case_cont.ped plink.assoc.logistic plink.eigenval plink.eigenvec plink.log root@PC1:/home/test# head -n 3 plink.assoc.logistic ## 查看结果 CHR SNP BP A1 TEST NMISS OR STAT P 1 snp1 3046 A ADD 288 1.069 0.237 0.8126 1 snp2 3092 T ADD 288 1.088 0.3019 0.7627
002、R
root@PC1:/home/test# ls gwas_case_cont.map gwas_case_cont.ped root@PC1:/home/test# plink --file gwas_case_cont --pca 3 1> /dev/null root@PC1:/home/test# ls gwas_case_cont.map gwas_case_cont.ped plink.eigenval plink.eigenvec plink.log root@PC1:/home/test# head -n 3 plink.eigenvec A1 A1 0.0161698 0.108569 -0.0182556 A2 A2 -0.0582408 0.00512212 0.0141237 A3 A3 -0.0363562 -0.0640895 0.0172107 root@PC1:/home/test# plink --file gwas_case_cont --recode A 1> /dev/null root@PC1:/home/test# ls gwas_case_cont.map gwas_case_cont.ped plink.eigenval plink.eigenvec plink.log plink.raw root@PC1:/home/test# head -n 5 plink.raw | cut -d " " -f 1-8 FID IID PAT MAT SEX PHENOTYPE snp1_A snp2_T A1 A1 0 0 1 1 1 1 A2 A2 0 0 1 1 0 0 A3 A3 0 0 1 1 0 0 A4 A4 0 0 1 1 0 0
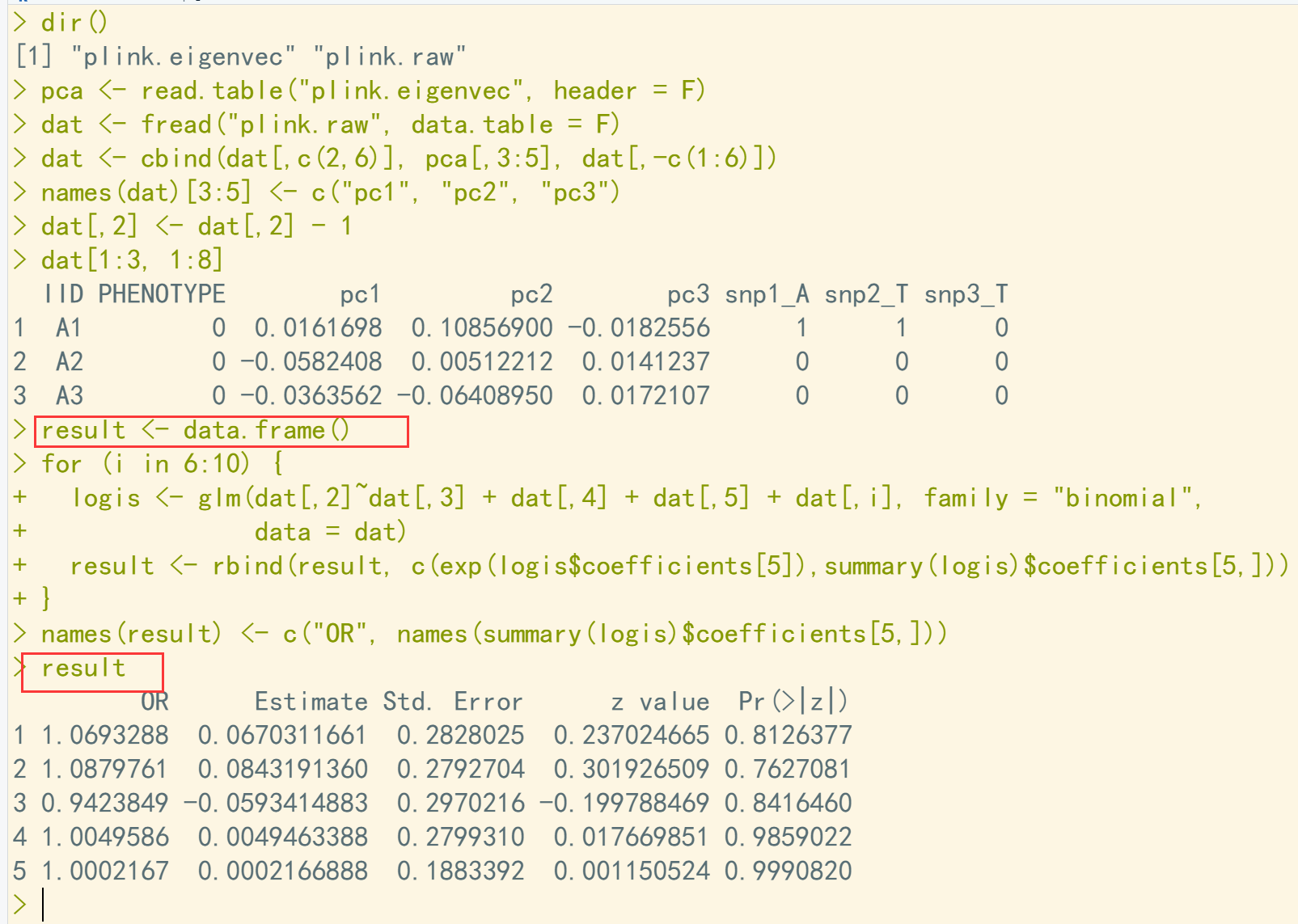
dir() pca <- read.table("plink.eigenvec", header = F) dat <- fread("plink.raw", data.table = F) dat <- cbind(dat[,c(2,6)], pca[,3:5], dat[,-c(1:6)]) names(dat)[3:5] <- c("pc1", "pc2", "pc3") dat[,2] <- dat[,2] - 1 dat[1:3, 1:8] result <- data.frame() for (i in 6:10) { logis <- glm(dat[,2]~dat[,3] + dat[,4] + dat[,5] + dat[,i], family = "binomial", data = dat) ## 逻辑回归,增加协变量, 同时计算OR,OR = exp(beta); se = beta/stat result <- rbind(result, c(exp(logis$coefficients[5]),summary(logis)$coefficients[5,])) } names(result) <- c("OR", names(summary(logis)$coefficients[5,])) result
plink验证:






【推荐】国内首个AI IDE,深度理解中文开发场景,立即下载体验Trae
【推荐】编程新体验,更懂你的AI,立即体验豆包MarsCode编程助手
【推荐】抖音旗下AI助手豆包,你的智能百科全书,全免费不限次数
【推荐】轻量又高性能的 SSH 工具 IShell:AI 加持,快人一步
· 震惊!C++程序真的从main开始吗?99%的程序员都答错了
· 【硬核科普】Trae如何「偷看」你的代码?零基础破解AI编程运行原理
· 单元测试从入门到精通
· 上周热点回顾(3.3-3.9)
· winform 绘制太阳,地球,月球 运作规律