利用基因型数据构建G矩阵
001、plink + sommer实现
root@PC1:/home/test# ls outcome.map outcome.ped root@PC1:/home/test# plink --file outcome --recode A 1> /dev/null ## 将A\T\C\G转换为0\1\2形式 root@PC1:/home/test# ls outcome.map outcome.ped plink.log plink.nosex plink.raw
library(sommer) library(data.table) dir() dat <- fread("plink.raw") g = as.matrix(dat[,!c(1:6)]) rownames(g) = dat$IID gmat = A.mat(g-1) gmat_cor <- gmat for (i in 1:nrow(gmat_cor)) { for (j in 1:ncol(gmat_cor)) { gmat_cor[i,j] <- gmat[i,j]/sqrt(gmat[i,i] * gmat[j,j]) } } dim(gmat_cor) gmat_cor[1:5,1:5] write.table(gmat_cor, "sommer_g.txt",row.names = T, col.names = T, sep = "\t", quote = F)
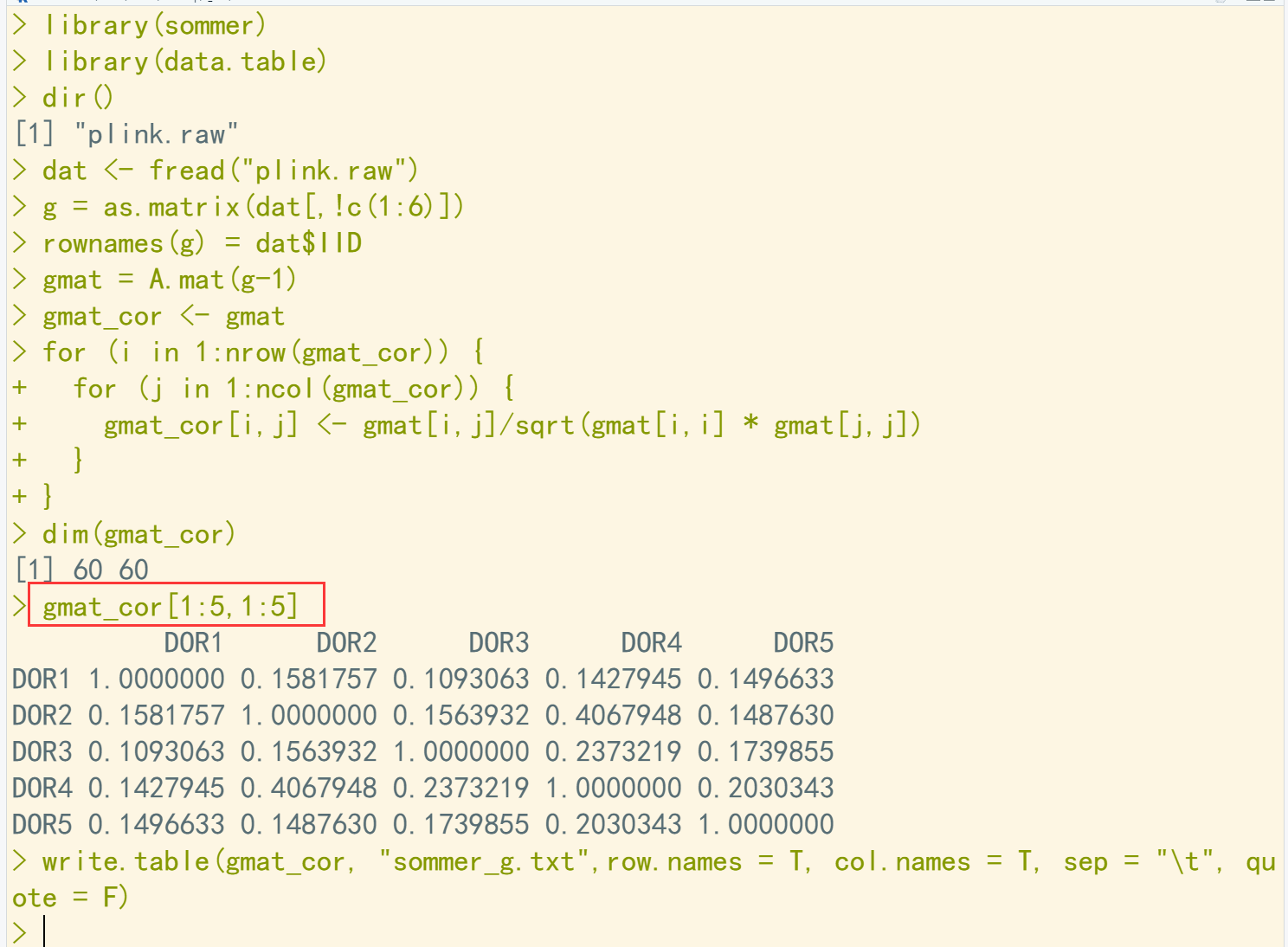
002、gemma实现
root@PC1:/home/test# ls outcome.map outcome.ped root@PC1:/home/test# plink --file outcome --make-bed 1> /dev/null ## 生成二进制文件 root@PC1:/home/test# ls outcome.map outcome.ped plink.bed plink.bim plink.fam plink.log root@PC1:/home/test# /home/software/gemma-0.98.5-linux-static-AMD64 -bfile plink -gk 2 -o gemma -outdir ./ ## 构建g矩阵 GEMMA 0.98.5 (2021-08-25) by Xiang Zhou, Pjotr Prins and team (C) 2012-2021 Reading Files ... ## number of total individuals = 60 ## number of analyzed individuals = 60 ## number of covariates = 1 ## number of phenotypes = 1 ## number of total SNPs/var = 42629 ## number of analyzed SNPs = 42629 Calculating Relatedness Matrix ... ================================================== 100% **** INFO: Done. root@PC1:/home/test# ls gemma.log.txt gemma.sXX.txt outcome.map outcome.ped plink.bed plink.bim plink.fam plink.log root@PC1:/home/test# head -n 5 gemma.sXX.txt | cut -f 1-5 0.9890465761 0.1073153377 0.1182919093 0.1263857559 0.1589786895 0.1073153377 0.9722035427 0.1886718834 0.2216020996 0.1799985087 0.1182919093 0.1886718834 0.9714687294 0.1556666191 0.1414708648 0.1263857559 0.2216020996 0.1556666191 0.9272709102 0.141052078 0.1589786895 0.1799985087 0.1414708648 0.141052078 0.9287239597
dir() sommer <- fread("sommer_g.txt") sommer[1:5, 1:5] gemma <- fread("gemma.sXX.txt") gemma[1:5, 1:5] cor(sommer$DOR1, gemma$V1) ## 计算相关系数
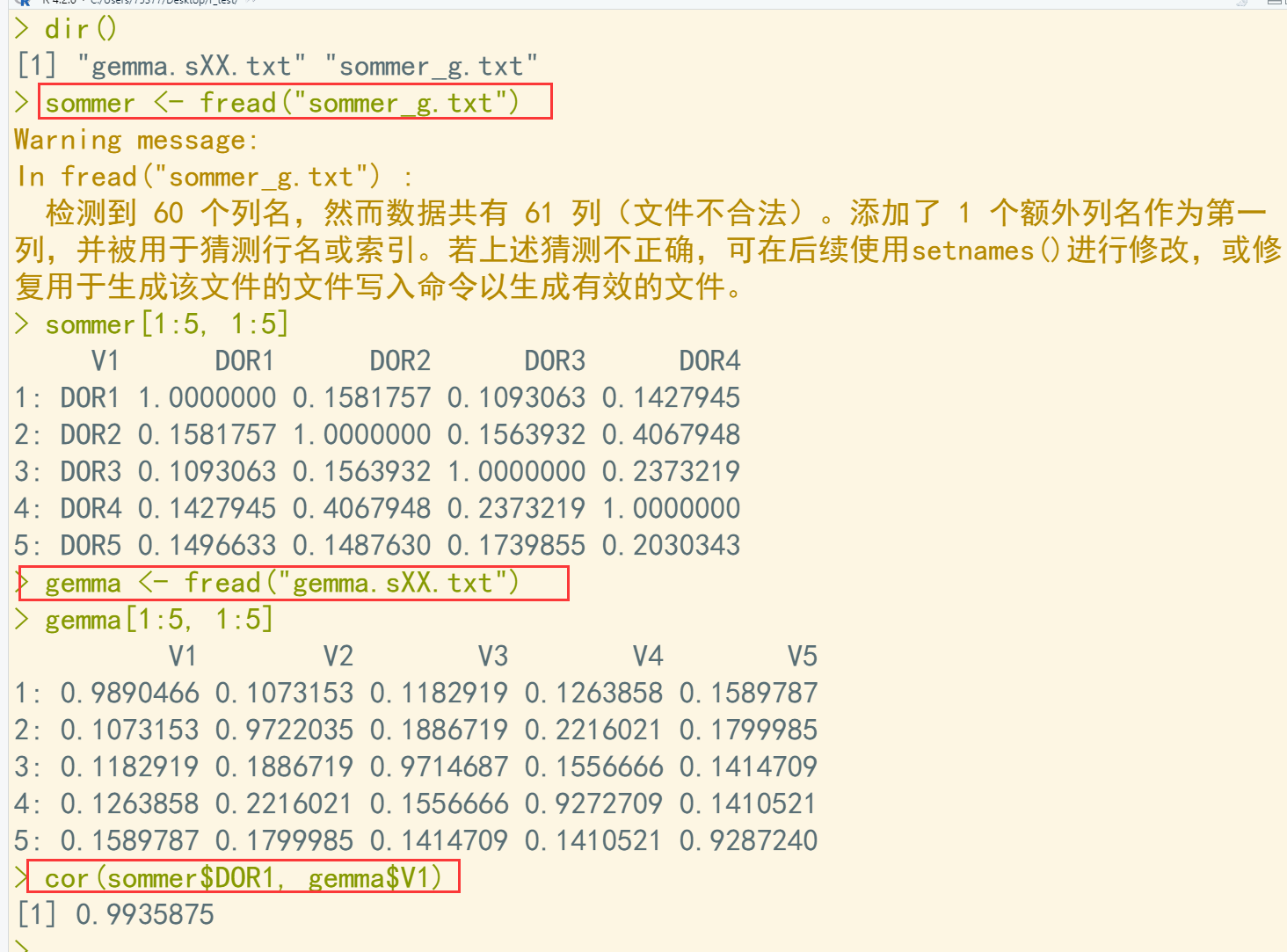
003、plink实现
root@PC1:/home/test# ls outcome.map outcome.ped root@PC1:/home/test# plink --file outcome --make-rel square 1> /dev/null ## plink生成G矩阵 root@PC1:/home/test# ls outcome.map outcome.ped plink.log plink.rel plink.rel.id root@PC1:/home/test# head -n 6 plink.rel | cut -f 1-6 1.04625 0.122221 0.132869 0.142839 0.178275 0.131377 0.122221 1.03097 0.209148 0.246329 0.201715 0.235886 0.132869 0.209148 1.02611 0.174476 0.160126 0.206312 0.142839 0.246329 0.174476 0.979224 0.16131 0.2332 0.178275 0.201715 0.160126 0.16131 0.98128 0.135685 0.131377 0.235886 0.206312 0.2332 0.135685 0.984521
dir() sommer <- fread("sommer_g.txt") sommer[1:3, 1:3] gemma <- fread("gemma.sXX.txt") gemma[1:3, 1:3] plink <- fread("plink.rel") plink[1:3, 1:3] cor(sommer$DOR1, plink$V1) cor(gemma$V1, plink$V1)

004、gcta实现
root@PC1:/home/test# ls outcome.map outcome.ped root@PC1:/home/test# plink --file outcome --make-bed 1> /dev/null ## 转二进制 root@PC1:/home/test# ls outcome.map outcome.ped plink.bed plink.bim plink.fam plink.log ## 构建矩阵 root@PC1:/home/test# /home/software/gcta_v1.94.0Beta_linux_kernel_3_x86_64/gcta_v1.94.0Beta_linux_kernel_3_x86_64_static --bfile plink --make-grm-gz --make-grm-alg 1 --out gcta 1> /dev/null root@PC1:/home/test# ls gcta.grm.gz gcta.grm.id gcta.log outcome.map outcome.ped plink.bed plink.bim plink.fam plink.log root@PC1:/home/test# gunzip gcta.grm. gzip: gcta.grm..gz: No such file or directory root@PC1:/home/test# gunzip gcta.grm.gz root@PC1:/home/test# ls gcta.grm gcta.grm.id gcta.log outcome.map outcome.ped plink.bed plink.bim plink.fam plink.log root@PC1:/home/test# head -n 3 gcta.grm ## 结果文件 1 1 1.593672e+04 1.068706e+00 2 1 1.593672e+04 1.294735e-01 2 2 1.593672e+04 1.078836e+00 root@PC1:/home/test# cut -f 1,2,4 gcta.grm > gcta.grm2 ## 修改格式 root@PC1:/home/test# ls gcta.grm gcta.grm2 gcta.grm.id gcta.log outcome.map outcome.ped plink.bed plink.bim plink.fam plink.log
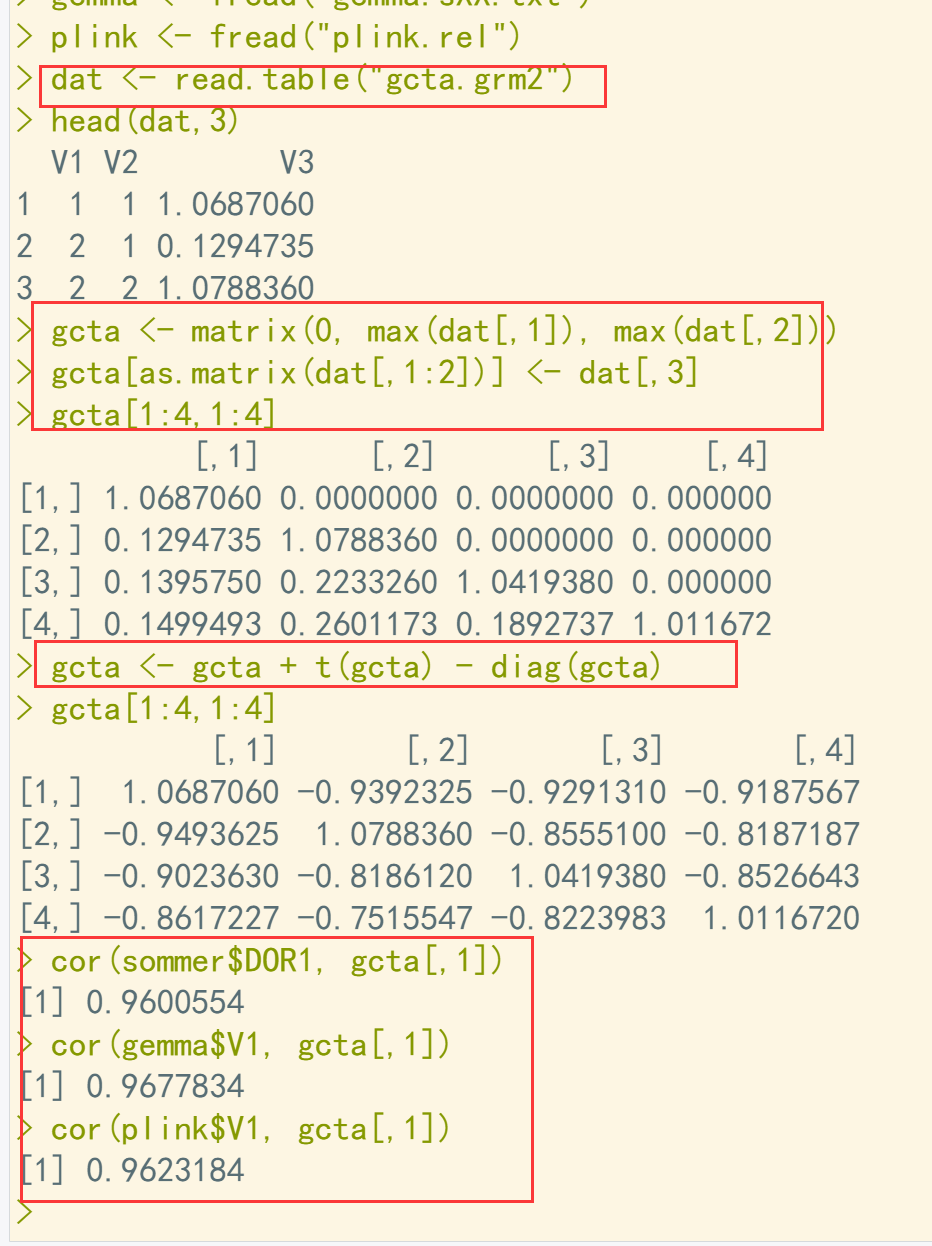
dir() sommer <- fread("sommer_g.txt") gemma <- fread("gemma.sXX.txt") plink <- fread("plink.rel") dat <- read.table("gcta.grm2") head(dat,3) gcta <- matrix(0, max(dat[,1]), max(dat[,2])) gcta[as.matrix(dat[,1:2])] <- dat[,3] ## 三元组转换为矩阵形式 gcta[1:4,1:4] gcta <- gcta + t(gcta) - diag(gcta) gcta[1:4,1:4] cor(sommer$DOR1, gcta[,1]) cor(gemma$V1, gcta[,1]) cor(plink$V1, gcta[,1])






【推荐】国内首个AI IDE,深度理解中文开发场景,立即下载体验Trae
【推荐】编程新体验,更懂你的AI,立即体验豆包MarsCode编程助手
【推荐】抖音旗下AI助手豆包,你的智能百科全书,全免费不限次数
【推荐】轻量又高性能的 SSH 工具 IShell:AI 加持,快人一步
· 震惊!C++程序真的从main开始吗?99%的程序员都答错了
· 【硬核科普】Trae如何「偷看」你的代码?零基础破解AI编程运行原理
· 单元测试从入门到精通
· 上周热点回顾(3.3-3.9)
· winform 绘制太阳,地球,月球 运作规律
2021-07-29 awk命令末尾数字的作用
2021-07-29 c语言中如何创建、存储、输出字符串、输出字符串的大小、字符串的长度