gemma 软件对数量性状 混合线性模型T值及P值的计算
001、
root@DESKTOP-1N42TVH:/home/test4# ls gwas_test.bed gwas_test.bim gwas_test.fam root@DESKTOP-1N42TVH:/home/test4# /home/software/gemma-0.98.5-linux-static-AMD64 -bfile gwas_test -gk -o kin 1> /dev/null **** INFO: Done. root@DESKTOP-1N42TVH:/home/test4# /home/software/gemma-0.98.5-linux-static-AMD64 -bfile gwas_test -k output/kin.cXX.txt -lmm -o result 1> /dev/null **** INFO: Done. root@DESKTOP-1N42TVH:/home/test4# ls gwas_test.bed gwas_test.bim gwas_test.fam output root@DESKTOP-1N42TVH:/home/test4# cd output/ root@DESKTOP-1N42TVH:/home/test4/output# ls kin.cXX.txt kin.log.txt result.assoc.txt result.log.txt root@DESKTOP-1N42TVH:/home/test4/output# head -n 3 result.assoc.txt chr rs ps n_miss allele1 allele0 af beta se logl_H1 l_remle p_wald 1 snp1 2802 0 G T 0.099 9.393582e-01 7.917546e+00 -3.077033e+03 2.285023e+01 9.056027e-01 1 snp2 2823 0 T C 0.063 5.449975e+00 9.371469e+00 -3.076887e+03 2.260596e+01 5.611132e-01 root@DESKTOP-1N42TVH:/home/test4/output# awk 'NR != 1 {print $(NF - 4)/$(NF - 3)}' result.assoc.txt > t.txt root@DESKTOP-1N42TVH:/home/test4/output# head -n 3 t.txt 0.118643 0.58155 0.985854 root@DESKTOP-1N42TVH:/home/test4/output# paste result.assoc.txt <(sed '1i t' t.txt ) > data.txt root@DESKTOP-1N42TVH:/home/test4/output# head data.txt chr rs ps n_miss allele1 allele0 af beta se logl_H1 l_remle p_wald t 1 snp1 2802 0 G T 0.099 9.393582e-01 7.917546e+00 -3.077033e+03 2.285023e+01 9.056027e-010.118643 1 snp2 2823 0 T C 0.063 5.449975e+00 9.371469e+00 -3.076887e+03 2.260596e+01 5.611132e-010.58155 1 snp3 4512 0 G A 0.067 9.014939e+00 9.144296e+00 -3.076572e+03 2.237338e+01 3.246472e-010.985854 1 snp4 16529 0 T C 0.055 -2.639543e+00 1.041215e+01 -3.076994e+03 2.293325e+01 7.999739e-01-0.253506 1 snp5 16578 0 G C 0.065 -2.507825e+00 9.000474e+00 -3.077046e+03 2.274653e+01 7.806337e-01-0.278633 1 snp6 16579 0 C A 0.275 1.898949e+00 5.196884e+00 -3.077006e+03 2.320171e+01 7.149551e-010.365401 1 snp7 16635 0 G C 0.088 4.344375e+00 8.115947e+00 -3.076915e+03 2.244365e+01 5.926710e-010.535289 1 snp8 20879 0 T G 0.130 3.405991e-01 6.905055e+00 -3.077041e+03 2.264882e+01 9.606777e-010.0493261 1 snp9 20908 0 C T 0.167 -1.752315e-01 6.246462e+00 -3.077057e+03 2.267830e+01 9.776303e-01-
002、R语言中验证
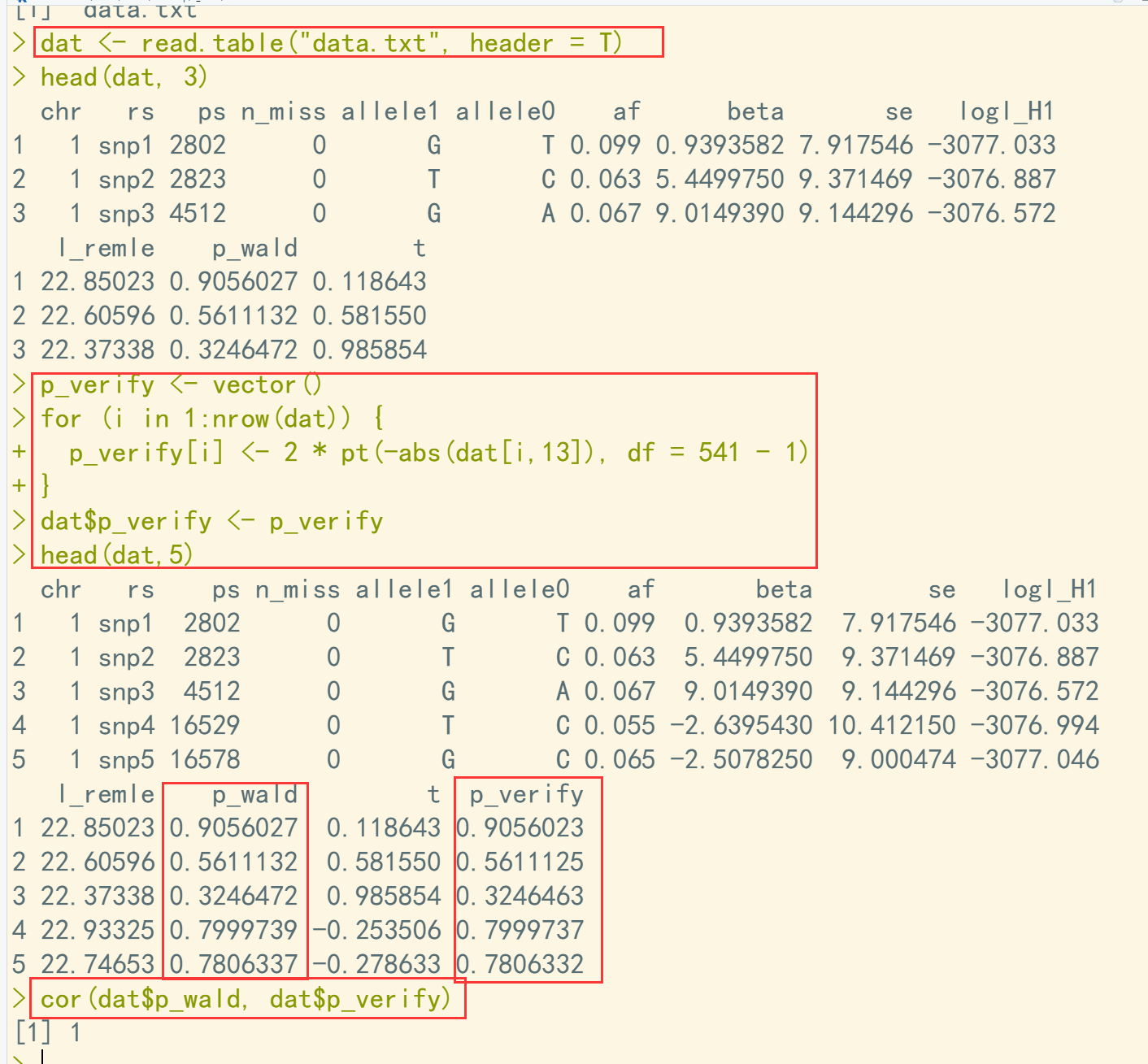
dir() dat <- read.table("data.txt", header = T) head(dat, 3) p_verify <- vector() for (i in 1:nrow(dat)) { p_verify[i] <- 2 * pt(-abs(dat[i,13]), df = 541 - 1) } dat$p_verify <- p_verify head(dat,5) cor(dat$p_wald, dat$p_verify)
分类:
生信






【推荐】国内首个AI IDE,深度理解中文开发场景,立即下载体验Trae
【推荐】编程新体验,更懂你的AI,立即体验豆包MarsCode编程助手
【推荐】抖音旗下AI助手豆包,你的智能百科全书,全免费不限次数
【推荐】轻量又高性能的 SSH 工具 IShell:AI 加持,快人一步
· 震惊!C++程序真的从main开始吗?99%的程序员都答错了
· 【硬核科普】Trae如何「偷看」你的代码?零基础破解AI编程运行原理
· 单元测试从入门到精通
· 上周热点回顾(3.3-3.9)
· winform 绘制太阳,地球,月球 运作规律
2021-07-15 4-4
2021-07-15 4-2-4
2021-07-15 4-1-3
2021-07-15 c语言中字符的声明和输出