plink 软件 --hardy参数计算哈代温伯格平衡中 p值的来源
基本公式:

001、计算卡方值x^2, 然后根据自由度计算p值。
root@DESKTOP-1N42TVH:/home/test6# ls result.map result.ped root@DESKTOP-1N42TVH:/home/test6# plink --file result --hardy --out hardy ## plink计算p值 PLINK v1.90b6.26 64-bit (2 Apr 2022) www.cog-genomics.org/plink/1.9/ (C) 2005-2022 Shaun Purcell, Christopher Chang GNU General Public License v3 Logging to hardy.log. Options in effect: --file result --hardy --out hardy 16007 MB RAM detected; reserving 8003 MB for main workspace. .ped scan complete (for binary autoconversion). Performing single-pass .bed write (442957 variants, 407 people). --file: hardy-temporary.bed + hardy-temporary.bim + hardy-temporary.fam written. 442957 variants loaded from .bim file. 407 people (0 males, 0 females, 407 ambiguous) loaded from .fam. Ambiguous sex IDs written to hardy.nosex . Using 1 thread (no multithreaded calculations invoked). Before main variant filters, 407 founders and 0 nonfounders present. Calculating allele frequencies... done. Total genotyping rate is exactly 1. --hardy: Writing Hardy-Weinberg report (founders only) to hardy.hwe ... done. root@DESKTOP-1N42TVH:/home/test6# ls hardy.hwe hardy.log hardy.nosex result.map result.ped root@DESKTOP-1N42TVH:/home/test6# plink --file result --freq --out freq ## 计算基因频率 PLINK v1.90b6.26 64-bit (2 Apr 2022) www.cog-genomics.org/plink/1.9/ (C) 2005-2022 Shaun Purcell, Christopher Chang GNU General Public License v3 Logging to freq.log. Options in effect: --file result --freq --out freq 16007 MB RAM detected; reserving 8003 MB for main workspace. .ped scan complete (for binary autoconversion). Performing single-pass .bed write (442957 variants, 407 people). --file: freq-temporary.bed + freq-temporary.bim + freq-temporary.fam written. 442957 variants loaded from .bim file. 407 people (0 males, 0 females, 407 ambiguous) loaded from .fam. Ambiguous sex IDs written to freq.nosex . Using 1 thread (no multithreaded calculations invoked). Before main variant filters, 407 founders and 0 nonfounders present. Calculating allele frequencies... done. Total genotyping rate is exactly 1. --freq: Allele frequencies (founders only) written to freq.frq . root@DESKTOP-1N42TVH:/home/test6# ls freq.frq freq.log freq.nosex hardy.hwe hardy.log hardy.nosex result.map result.ped
合并p值文件和基因频率文件:
root@DESKTOP-1N42TVH:/home/test6# ls freq.frq freq.log freq.nosex hardy.hwe hardy.log hardy.nosex result.map result.ped root@DESKTOP-1N42TVH:/home/test6# head hardy.hwe CHR SNP TEST A1 A2 GENO O(HET) E(HET) P 1 oar3_OAR1_17218 ALL(NP) G A 9/105/293 0.258 0.2565 1 1 oar3_OAR1_20658 ALL(NP) C A 25/123/259 0.3022 0.3347 0.05401 1 oar3_OAR1_28296 ALL(NP) A G 17/140/250 0.344 0.3361 0.7679 1 oar3_OAR1_31152 ALL(NP) G A 103/185/119 0.4545 0.4992 0.07405 1 oar3_OAR1_38175 ALL(NP) A G 14/119/274 0.2924 0.296 0.8667 1 oar3_OAR1_38264 ALL(NP) A G 39/191/177 0.4693 0.4425 0.2626 1 s64199.1 ALL(NP) A G 6/101/300 0.2482 0.2391 0.5385 1 oar3_OAR1_52919 ALL(NP) G A 98/198/111 0.4865 0.4995 0.62 1 oar3_OAR1_55363 ALL(NP) A G 70/166/171 0.4079 0.4692 0.00837 root@DESKTOP-1N42TVH:/home/test6# head freq.frq CHR SNP A1 A2 MAF NCHROBS 1 oar3_OAR1_17218 G A 0.1511 814 1 oar3_OAR1_20658 C A 0.2125 814 1 oar3_OAR1_28296 A G 0.2138 814 1 oar3_OAR1_31152 G A 0.4803 814 1 oar3_OAR1_38175 A G 0.1806 814 1 oar3_OAR1_38264 A G 0.3305 814 1 s64199.1 A G 0.1388 814 1 oar3_OAR1_52919 G A 0.484 814 1 oar3_OAR1_55363 A G 0.3759 814 root@DESKTOP-1N42TVH:/home/test6# paste hardy.hwe <(awk '{print $5}' freq.frq ) | head CHR SNP TEST A1 A2 GENO O(HET) E(HET) P MAF 1 oar3_OAR1_17218 ALL(NP) G A 9/105/293 0.258 0.2565 1 0.1511 1 oar3_OAR1_20658 ALL(NP) C A 25/123/259 0.3022 0.3347 0.05401 0.2125 1 oar3_OAR1_28296 ALL(NP) A G 17/140/250 0.344 0.3361 0.7679 0.2138 1 oar3_OAR1_31152 ALL(NP) G A 103/185/119 0.4545 0.4992 0.07405 0.4803 1 oar3_OAR1_38175 ALL(NP) A G 14/119/274 0.2924 0.296 0.8667 0.1806 1 oar3_OAR1_38264 ALL(NP) A G 39/191/177 0.4693 0.4425 0.2626 0.3305 1 s64199.1 ALL(NP) A G 6/101/300 0.2482 0.2391 0.5385 0.1388 1 oar3_OAR1_52919 ALL(NP) G A 98/198/111 0.4865 0.4995 0.62 0.484 1 oar3_OAR1_55363 ALL(NP) A G 70/166/171 0.4079 0.4692 0.00837 0.3759 root@DESKTOP-1N42TVH:/home/test6# paste hardy.hwe <(awk '{print $5}' freq.frq ) | sed 's/^[\t ]\+//g' | sed 's/[\t ]\+/\t/g' | head CHR SNP TEST A1 A2 GENO O(HET) E(HET) P MAF 1 oar3_OAR1_17218 ALL(NP) G A 9/105/293 0.258 0.2565 1 0.1511 1 oar3_OAR1_20658 ALL(NP) C A 25/123/259 0.3022 0.3347 0.05401 0.2125 1 oar3_OAR1_28296 ALL(NP) A G 17/140/250 0.344 0.3361 0.7679 0.2138 1 oar3_OAR1_31152 ALL(NP) G A 103/185/119 0.4545 0.4992 0.07405 0.4803 1 oar3_OAR1_38175 ALL(NP) A G 14/119/274 0.2924 0.296 0.8667 0.1806 1 oar3_OAR1_38264 ALL(NP) A G 39/191/177 0.4693 0.4425 0.2626 0.3305 1 s64199.1 ALL(NP) A G 6/101/300 0.2482 0.2391 0.5385 0.1388 1 oar3_OAR1_52919 ALL(NP) G A 98/198/111 0.4865 0.4995 0.62 0.484 1 oar3_OAR1_55363 ALL(NP) A G 70/166/171 0.4079 0.4692 0.00837 0.3759 root@DESKTOP-1N42TVH:/home/test6# paste hardy.hwe <(awk '{print $5}' freq.frq ) | sed 's/^[\t ]\+//g' | sed 's/[\t ]\+/\t/g' | awk '{OFS = "\t"; print $0, $NF * $NF * 407, 2 * $NF * (1 - $NF) * 407, (1 - $NF) * (1 - $NF) * 407}' | head CHR SNP TEST A1 A2 GENO O(HET) E(HET) P MAF 0 0 407 1 oar3_OAR1_17218 ALL(NP) G A 9/105/293 0.258 0.2565 1 0.1511 9.2923 104.411 293.297 1 oar3_OAR1_20658 ALL(NP) C A 25/123/259 0.3022 0.3347 0.05401 0.2125 18.3786 136.218 252.404 1 oar3_OAR1_28296 ALL(NP) A G 17/140/250 0.344 0.3361 0.7679 0.2138 18.6041 136.825 251.571 1 oar3_OAR1_31152 ALL(NP) G A 103/185/119 0.4545 0.4992 0.07405 0.4803 93.8901 203.184 109.926 1 oar3_OAR1_38175 ALL(NP) A G 14/119/274 0.2924 0.296 0.8667 0.1806 13.2749 120.459 273.266 1 oar3_OAR1_38264 ALL(NP) A G 39/191/177 0.4693 0.4425 0.2626 0.3305 44.4567 180.114 182.43 1 s64199.1 ALL(NP) A G 6/101/300 0.2482 0.2391 0.5385 0.1388 7.84103 97.3011 301.858 1 oar3_OAR1_52919 ALL(NP) G A 98/198/111 0.4865 0.4995 0.62 0.484 95.3422 203.292 108.366 1 oar3_OAR1_55363 ALL(NP) A G 70/166/171 0.4079 0.4692 0.00837 0.3759 57.5094 190.964 158.527
root@DESKTOP-1N42TVH:/home/test6# paste hardy.hwe <(awk '{print $5}' freq.frq ) | sed 's/^[\t ]\+//g' | sed 's/[\t ]\+/\t/g' | awk '{OFS = "\t"; print $0, $NF * $NF * 407, 2 * $NF * (1 - $NF) * 407, (1 - $NF) * (1 - $NF) * 407}' | awk '{print $6, $(NF - 2) , $(NF - 1) , $NF}' | head GENO 0 0 407 9/105/293 9.2923 104.411 293.297 25/123/259 18.3786 136.218 252.404 17/140/250 18.6041 136.825 251.571 103/185/119 93.8901 203.184 109.926 14/119/274 13.2749 120.459 273.266 39/191/177 44.4567 180.114 182.43 6/101/300 7.84103 97.3011 301.858 98/198/111 95.3422 203.292 108.366 70/166/171 57.5094 190.964 158.527 root@DESKTOP-1N42TVH:/home/test6# paste hardy.hwe <(awk '{print $5}' freq.frq ) | sed 's/^[\t ]\+//g' | sed 's/[\t ]\+/\t/g' | awk '{OFS = "\t"; print $0, $NF * $NF * 407, 2 * $NF * (1 - $NF) * 407, (1 - $NF) * (1 - $NF) * 407}' | awk '{print $6, $(NF - 2) , $(NF - 1) , $NF}' | sed 1d | sed 's/\// /g' | head 9 105 293 9.2923 104.411 293.297 25 123 259 18.3786 136.218 252.404 17 140 250 18.6041 136.825 251.571 103 185 119 93.8901 203.184 109.926 14 119 274 13.2749 120.459 273.266 39 191 177 44.4567 180.114 182.43 6 101 300 7.84103 97.3011 301.858 98 198 111 95.3422 203.292 108.366 70 166 171 57.5094 190.964 158.527 59 184 164 56.0199 189.954 161.026 root@DESKTOP-1N42TVH:/home/test6# paste hardy.hwe <(awk '{print $5}' freq.frq ) | sed 's/^[\t ]\+//g' | sed 's/[\t ]\+/\t/g' | awk '{OFS = "\t"; print $0, $NF * $NF * 407, 2 * $NF * (1 - $NF) * 407, (1 - $NF) * (1 - $NF) * 407}' | awk '{print $6, $(NF - 2) , $(NF - 1) , $NF}' | sed 1d | sed 's/\// /g' | sed 's/ /\t/g' | head 9 105 293 9.2923 104.411 293.297 25 123 259 18.3786 136.218 252.404 17 140 250 18.6041 136.825 251.571 103 185 119 93.8901 203.184 109.926 14 119 274 13.2749 120.459 273.266 39 191 177 44.4567 180.114 182.43 6 101 300 7.84103 97.3011 301.858 98 198 111 95.3422 203.292 108.366 70 166 171 57.5094 190.964 158.527 59 184 164 56.0199 189.954 161.026 root@DESKTOP-1N42TVH:/home/test6# paste hardy.hwe <(awk '{print $5}' freq.frq ) | sed 's/^[\t ]\+//g' | sed 's/[\t ]\+/\t/g' | awk '{OFS = "\t"; print $0, $NF * $NF * 407, 2 * $NF * (1 - $NF) * 407, (1 - $NF) * (1 - $NF) * 407}' | awk '{print $6, $(NF - 2) , $(NF - 1) , $NF}' | sed 1d | sed 's/\// /g' | sed 's/ /\t/g' > com.txt
计算卡方值x^2:
root@DESKTOP-1N42TVH:/home/test6# head com.txt 9 105 293 9.2923 104.411 293.297 25 123 259 18.3786 136.218 252.404 17 140 250 18.6041 136.825 251.571 103 185 119 93.8901 203.184 109.926 14 119 274 13.2749 120.459 273.266 39 191 177 44.4567 180.114 182.43 6 101 300 7.84103 97.3011 301.858 98 198 111 95.3422 203.292 108.366 70 166 171 57.5094 190.964 158.527 59 184 164 56.0199 189.954 161.026 root@DESKTOP-1N42TVH:/home/test6# awk '{print ($1 - $4)*($1 - $4)/$4 + ($2 - $5)*($2 - $5)/$5 + ($3 - $6)*($3 - $6)/$6}' com.txt > x_sqrare.txt root@DESKTOP-1N42TVH:/home/test6# head x_sqrare.txt 0.012818 3.84053 0.221796 3.26032 0.0592493 1.48933 0.584314 0.275872 6.95769 0.400085
利用R计算P值:
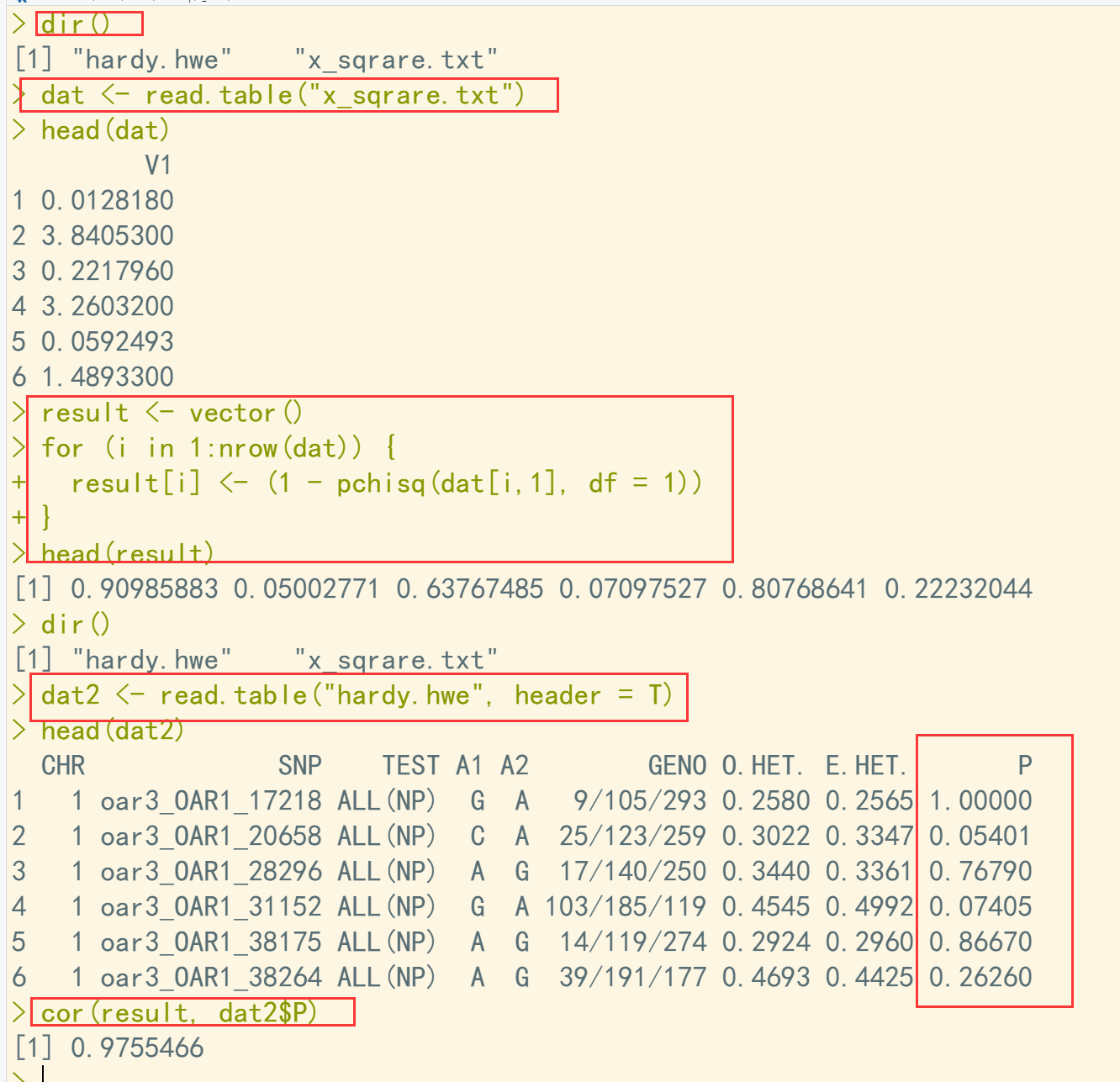
dir() dat <- read.table("x_sqrare.txt") head(dat) result <- vector() for (i in 1:nrow(dat)) { result[i] <- (1 - pchisq(dat[i,1], df = 1)) } head(result) ## 结果不完全一致,相关系数0.97??? dir() dat2 <- read.table("hardy.hwe", header = T) head(dat2) cor(result, dat2$P)






【推荐】国内首个AI IDE,深度理解中文开发场景,立即下载体验Trae
【推荐】编程新体验,更懂你的AI,立即体验豆包MarsCode编程助手
【推荐】抖音旗下AI助手豆包,你的智能百科全书,全免费不限次数
【推荐】轻量又高性能的 SSH 工具 IShell:AI 加持,快人一步
· 震惊!C++程序真的从main开始吗?99%的程序员都答错了
· 【硬核科普】Trae如何「偷看」你的代码?零基础破解AI编程运行原理
· 单元测试从入门到精通
· 上周热点回顾(3.3-3.9)
· winform 绘制太阳,地球,月球 运作规律
2021-07-13 R语言返回重复的向量以重复向量的索引
2021-07-13 c语言中的整数溢出问题