plink 软件中 --het参数
001、
root@DESKTOP-1N42TVH:/home/test4# ls outcome.map outcome.ped root@DESKTOP-1N42TVH:/home/test4# plink --file outcome --het --out test PLINK v1.90b6.26 64-bit (2 Apr 2022) www.cog-genomics.org/plink/1.9/ (C) 2005-2022 Shaun Purcell, Christopher Chang GNU General Public License v3 Logging to test.log. Options in effect: --file outcome --het --out test 16007 MB RAM detected; reserving 8003 MB for main workspace. .ped scan complete (for binary autoconversion). Performing single-pass .bed write (21542 variants, 20 people). --file: test-temporary.bed + test-temporary.bim + test-temporary.fam written. 21542 variants loaded from .bim file. 20 people (0 males, 0 females, 20 ambiguous) loaded from .fam. Ambiguous sex IDs written to test.nosex . Using 1 thread (no multithreaded calculations invoked). Before main variant filters, 20 founders and 0 nonfounders present. Calculating allele frequencies... done. Total genotyping rate is exactly 1. 21542 variants and 20 people pass filters and QC. Note: No phenotypes present. --het: 19263 variants scanned, report written to test.het . root@DESKTOP-1N42TVH:/home/test4# ls outcome.map outcome.ped test.het test.log test.nosex root@DESKTOP-1N42TVH:/home/test4# cat test.het FID IID O(HOM) E(HOM) N(NM) F DOR 1 12374 1.257e+04 19263 -0.02885 DOR 2 13047 1.257e+04 19263 0.07166 DOR 3 11857 1.257e+04 19263 -0.1061 DOR 4 12037 1.257e+04 19263 -0.07918 DOR 5 12558 1.257e+04 19263 -0.00137 DOR 6 12152 1.257e+04 19263 -0.062 DOR 7 11860 1.257e+04 19263 -0.1056 DOR 9 12607 1.257e+04 19263 0.005948 DOR 10 11988 1.257e+04 19263 -0.0865 DOR 11 12692 1.257e+04 19263 0.01864 DOR 12 11958 1.257e+04 19263 -0.09098 DOR 13 12396 1.257e+04 19263 -0.02556 DOR 14 12080 1.257e+04 19263 -0.07276 DOR 15 11938 1.257e+04 19263 -0.09396 DOR 16 12414 1.257e+04 19263 -0.02288 DOR 17 11979 1.257e+04 19263 -0.08784 DOR 18 12288 1.257e+04 19263 -0.04169 DOR 19 12247 1.257e+04 19263 -0.04782 DOR 20 12790 1.257e+04 19263 0.03328 DOR 22 12334 1.257e+04 19263 -0.03482
002、F来源:https://www.cog-genomics.org/plink/1.9/basic_stats#ibc

003、验证:
root@DESKTOP-1N42TVH:/home/test4# ls outcome.map outcome.ped test.het test.log test.nosex root@DESKTOP-1N42TVH:/home/test4# cat test.het FID IID O(HOM) E(HOM) N(NM) F DOR 1 12374 1.257e+04 19263 -0.02885 DOR 2 13047 1.257e+04 19263 0.07166 DOR 3 11857 1.257e+04 19263 -0.1061 DOR 4 12037 1.257e+04 19263 -0.07918 DOR 5 12558 1.257e+04 19263 -0.00137 DOR 6 12152 1.257e+04 19263 -0.062 DOR 7 11860 1.257e+04 19263 -0.1056 DOR 9 12607 1.257e+04 19263 0.005948 DOR 10 11988 1.257e+04 19263 -0.0865 DOR 11 12692 1.257e+04 19263 0.01864 DOR 12 11958 1.257e+04 19263 -0.09098 DOR 13 12396 1.257e+04 19263 -0.02556 DOR 14 12080 1.257e+04 19263 -0.07276 DOR 15 11938 1.257e+04 19263 -0.09396 DOR 16 12414 1.257e+04 19263 -0.02288 DOR 17 11979 1.257e+04 19263 -0.08784 DOR 18 12288 1.257e+04 19263 -0.04169 DOR 19 12247 1.257e+04 19263 -0.04782 DOR 20 12790 1.257e+04 19263 0.03328 DOR 22 12334 1.257e+04 19263 -0.03482 root@DESKTOP-1N42TVH:/home/test4# awk 'NR != 1{print ($3 - $4)/($5 - $4)}' test.het -0.0292843 0.0712685 -0.106529 -0.0796354 -0.00179292 -0.0624533 -0.106081 0.00552816 -0.0869565 0.018228 -0.0914388 -0.0259973 -0.0732108 -0.094427 -0.0233079 -0.0883012 -0.0421336 -0.0482594 0.0328702 -0.0352607
004、 O(HOM)、E(HOM)、N(NM)如何计算
root@DESKTOP-1N42TVH:/home/test5# ls verify.map verify.ped root@DESKTOP-1N42TVH:/home/test5# cat verify.map 1 s64199.1 0 55910 1 OAR19_64675012.1 0 85204 1 OAR19_64715327.1 0 122948 1 OAR19_64803054.1 0 203750 1 DU281551_498.1 0 312707 1 s18939.1 0 356863 1 OAR1_88143.1 0 400518 1 s09912.1 0 487423 root@DESKTOP-1N42TVH:/home/test5# cat verify.ped DOR 1 0 0 0 -9 G G C C T G G G A G A C G G G C DOR 2 0 0 0 -9 C C G C T G G G G G A C A G C C DOR 3 0 0 0 -9 G G C C T T G G G G A A A G G C DOR 4 0 0 0 -9 G G C C G G G G G G A A G G G G DOR 5 0 0 0 -9 G G C C G G G G G G A A A G G C DOR 6 0 0 0 -9 G G C C G G G G G G A A A A C C DOR 7 0 0 0 -9 G G C C G G A G A A A A G G C C DOR 9 0 0 0 -9 G G C C G G A G A A A A G G C C DOR 10 0 0 0 -9 G G G C G G G G G G A A A G C C DOR 11 0 0 0 -9 G G C C G G G G A G C C A G G C root@DESKTOP-1N42TVH:/home/test5# plink --file verify --het --out test PLINK v1.90b6.26 64-bit (2 Apr 2022) www.cog-genomics.org/plink/1.9/ (C) 2005-2022 Shaun Purcell, Christopher Chang GNU General Public License v3 Logging to test.log. Options in effect: --file verify --het --out test 16007 MB RAM detected; reserving 8003 MB for main workspace. .ped scan complete (for binary autoconversion). Performing single-pass .bed write (8 variants, 10 people). --file: test-temporary.bed + test-temporary.bim + test-temporary.fam written. 8 variants loaded from .bim file. 10 people (0 males, 0 females, 10 ambiguous) loaded from .fam. Ambiguous sex IDs written to test.nosex . Using 1 thread (no multithreaded calculations invoked). Before main variant filters, 10 founders and 0 nonfounders present. Calculating allele frequencies... done. Total genotyping rate is exactly 1. 8 variants and 10 people pass filters and QC. Note: No phenotypes present. --het: 8 variants scanned, report written to test.het . root@DESKTOP-1N42TVH:/home/test5# ls test.het test.log test.nosex verify.map verify.ped
root@DESKTOP-1N42TVH:/home/test5# ls test.het test.log test.nosex verify.map verify.ped root@DESKTOP-1N42TVH:/home/test5# cat test.het FID IID O(HOM) E(HOM) N(NM) F DOR 1 4 5.525 8 -0.6162 DOR 2 4 5.525 8 -0.6162 DOR 3 6 5.525 8 0.1919 DOR 4 8 5.525 8 1 DOR 5 6 5.525 8 0.1919 DOR 6 8 5.525 8 1 DOR 7 7 5.525 8 0.596 DOR 9 7 5.525 8 0.596 DOR 10 6 5.525 8 0.1919 DOR 11 5 5.525 8 -0.2121 root@DESKTOP-1N42TVH:/home/test5# cat verify.ped DOR 1 0 0 0 -9 G G C C T G G G A G A C G G G C DOR 2 0 0 0 -9 C C G C T G G G G G A C A G C C DOR 3 0 0 0 -9 G G C C T T G G G G A A A G G C DOR 4 0 0 0 -9 G G C C G G G G G G A A G G G G DOR 5 0 0 0 -9 G G C C G G G G G G A A A G G C DOR 6 0 0 0 -9 G G C C G G G G G G A A A A C C DOR 7 0 0 0 -9 G G C C G G A G A A A A G G C C DOR 9 0 0 0 -9 G G C C G G A G A A A A G G C C DOR 10 0 0 0 -9 G G G C G G G G G G A A A G C C DOR 11 0 0 0 -9 G G C C G G G G A G C C A G G C
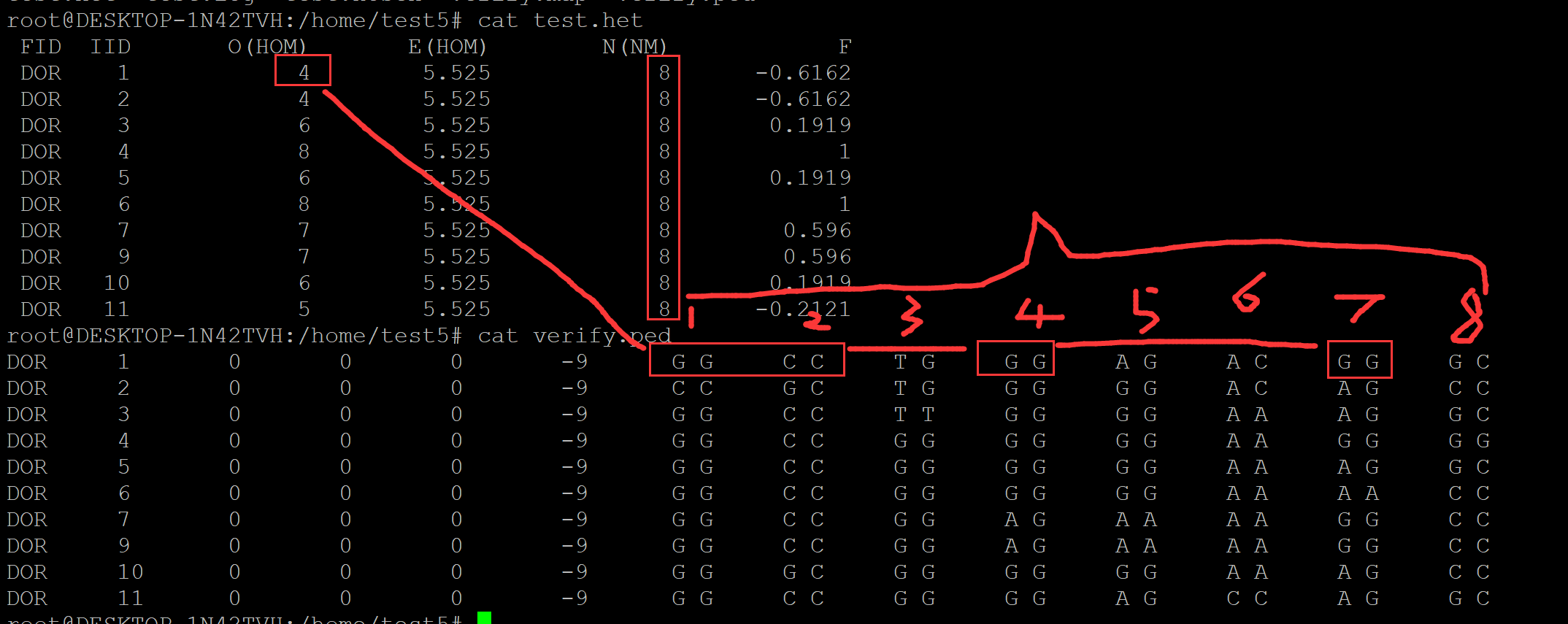
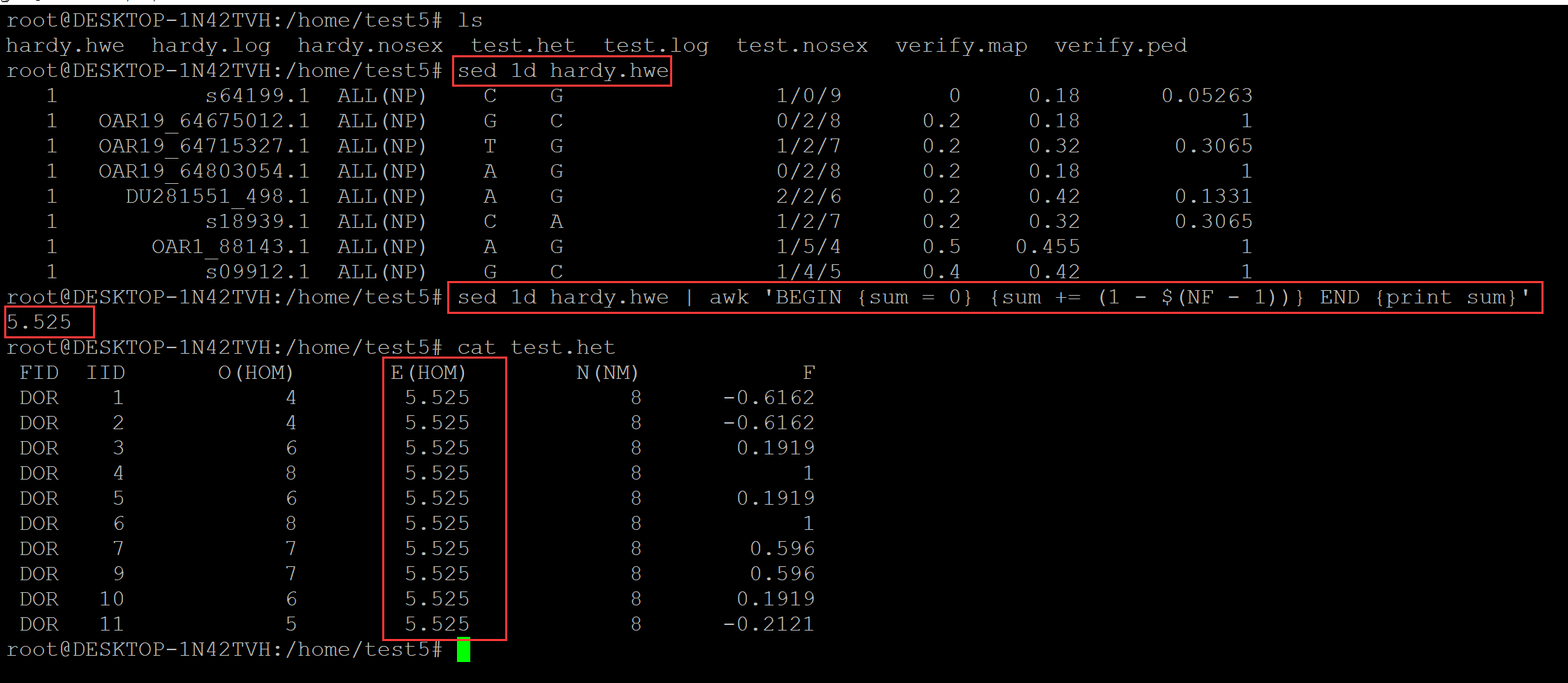
个体期待纯合度:E(HOM) 如何计算?
root@DESKTOP-1N42TVH:/home/test5# ls test.het test.log test.nosex verify.map verify.ped root@DESKTOP-1N42TVH:/home/test5# plink --file verify --hardy --out hardy PLINK v1.90b6.26 64-bit (2 Apr 2022) www.cog-genomics.org/plink/1.9/ (C) 2005-2022 Shaun Purcell, Christopher Chang GNU General Public License v3 Logging to hardy.log. Options in effect: --file verify --hardy --out hardy 16007 MB RAM detected; reserving 8003 MB for main workspace. .ped scan complete (for binary autoconversion). Performing single-pass .bed write (8 variants, 10 people). --file: hardy-temporary.bed + hardy-temporary.bim + hardy-temporary.fam written. 8 variants loaded from .bim file. 10 people (0 males, 0 females, 10 ambiguous) loaded from .fam. Ambiguous sex IDs written to hardy.nosex . Using 1 thread (no multithreaded calculations invoked). Before main variant filters, 10 founders and 0 nonfounders present. Calculating allele frequencies... done. Total genotyping rate is exactly 1. --hardy: Writing Hardy-Weinberg report (founders only) to hardy.hwe ... done. root@DESKTOP-1N42TVH:/home/test5# ls hardy.hwe hardy.log hardy.nosex test.het test.log test.nosex verify.map verify.ped root@DESKTOP-1N42TVH:/home/test5# cat hardy.hwe ## 倒数第二列,位点的期待杂合度, 由此可计算期待纯合度,所有位点期待纯合度累加, 获取个体期待纯合度 CHR SNP TEST A1 A2 GENO O(HET) E(HET) P 1 s64199.1 ALL(NP) C G 1/0/9 0 0.18 0.05263 1 OAR19_64675012.1 ALL(NP) G C 0/2/8 0.2 0.18 1 1 OAR19_64715327.1 ALL(NP) T G 1/2/7 0.2 0.32 0.3065 1 OAR19_64803054.1 ALL(NP) A G 0/2/8 0.2 0.18 1 1 DU281551_498.1 ALL(NP) A G 2/2/6 0.2 0.42 0.1331 1 s18939.1 ALL(NP) C A 1/2/7 0.2 0.32 0.3065 1 OAR1_88143.1 ALL(NP) A G 1/5/4 0.5 0.455 1 1 s09912.1 ALL(NP) G C 1/4/5 0.4 0.42 1 root@DESKTOP-1N42TVH:/home/test5# ls hardy.hwe hardy.log hardy.nosex test.het test.log test.nosex verify.map verify.ped root@DESKTOP-1N42TVH:/home/test5# cat hardy.hwe CHR SNP TEST A1 A2 GENO O(HET) E(HET) P 1 s64199.1 ALL(NP) C G 1/0/9 0 0.18 0.05263 1 OAR19_64675012.1 ALL(NP) G C 0/2/8 0.2 0.18 1 1 OAR19_64715327.1 ALL(NP) T G 1/2/7 0.2 0.32 0.3065 1 OAR19_64803054.1 ALL(NP) A G 0/2/8 0.2 0.18 1 1 DU281551_498.1 ALL(NP) A G 2/2/6 0.2 0.42 0.1331 1 s18939.1 ALL(NP) C A 1/2/7 0.2 0.32 0.3065 1 OAR1_88143.1 ALL(NP) A G 1/5/4 0.5 0.455 1 1 s09912.1 ALL(NP) G C 1/4/5 0.4 0.42 1 root@DESKTOP-1N42TVH:/home/test5# sed 1d hardy.hwe 1 s64199.1 ALL(NP) C G 1/0/9 0 0.18 0.05263 1 OAR19_64675012.1 ALL(NP) G C 0/2/8 0.2 0.18 1 1 OAR19_64715327.1 ALL(NP) T G 1/2/7 0.2 0.32 0.3065 1 OAR19_64803054.1 ALL(NP) A G 0/2/8 0.2 0.18 1 1 DU281551_498.1 ALL(NP) A G 2/2/6 0.2 0.42 0.1331 1 s18939.1 ALL(NP) C A 1/2/7 0.2 0.32 0.3065 1 OAR1_88143.1 ALL(NP) A G 1/5/4 0.5 0.455 1 1 s09912.1 ALL(NP) G C 1/4/5 0.4 0.42 1 root@DESKTOP-1N42TVH:/home/test5# sed 1d hardy.hwe | awk 'BEGIN {sum = 0} {sum += (1 - $(NF - 1))} END {print sum}' 5.525
分类:
生信






【推荐】国内首个AI IDE,深度理解中文开发场景,立即下载体验Trae
【推荐】编程新体验,更懂你的AI,立即体验豆包MarsCode编程助手
【推荐】抖音旗下AI助手豆包,你的智能百科全书,全免费不限次数
【推荐】轻量又高性能的 SSH 工具 IShell:AI 加持,快人一步
· 震惊!C++程序真的从main开始吗?99%的程序员都答错了
· 【硬核科普】Trae如何「偷看」你的代码?零基础破解AI编程运行原理
· 单元测试从入门到精通
· 上周热点回顾(3.3-3.9)
· winform 绘制太阳,地球,月球 运作规律
2021-07-12 c语言为什么设计这么多种数据类型
2021-07-12 c语言中不同进制数的表示
2021-07-12 计算机的字长是指什么