Seurat单细胞测序中高变异基因的选择程序验证
数据来源:https://www.jianshu.com/p/4f7aeae81ef1
1、
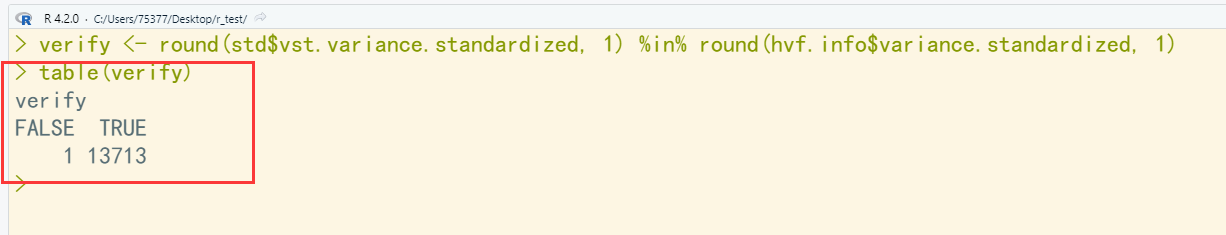
library(dplyr) library(Seurat) library(patchwork) # Load the PBMC dataset pbmc.data <- Read10X(data.dir = "C:/Users/75377/Desktop/r_test/hg19") # Initialize the Seurat object with the raw (non-normalized data). pbmc <- CreateSeuratObject(counts = pbmc.data, project = "pbmc3k", min.cells = 3, min.features = 200) pbmc ## An object of class Seurat ## 13714 features across 2700 samples within 1 assay ## Active assay: RNA (13714 features, 0 variable features) # The [[ operator can add columns to object metadata. This is a great place to stash QC stats pbmc[["percent.mt"]] <- PercentageFeatureSet(pbmc, pattern = "^MT-") # Visualize QC metrics as a violin plot VlnPlot(pbmc, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3) # FeatureScatter is typically used to visualize feature-feature relationships, but can be used # for anything calculated by the object, i.e. columns in object metadata, PC scores etc. plot1 <- FeatureScatter(pbmc, feature1 = "nCount_RNA", feature2 = "percent.mt") plot2 <- FeatureScatter(pbmc, feature1 = "nCount_RNA", feature2 = "nFeature_RNA") plot1 + plot2 pbmc <- subset(pbmc, subset = nFeature_RNA > 200 & nFeature_RNA < 2500 & percent.mt < 5) pbmc <- NormalizeData(pbmc) pbmc <- FindVariableFeatures(pbmc, selection.method = "vst", nfeatures = 2000) # Identify the 10 most highly variable genes top10 <- head(VariableFeatures(pbmc), 10) # plot variable features with and without labels plot1 <- VariableFeaturePlot(pbmc) plot2 <- LabelPoints(plot = plot1, points = top10, repel = TRUE) plot1 + plot2 std <- pbmc[["RNA"]]@meta.features std <- as.data.frame(std) fwrite(std, "std.txt", row.names = T, col.names = T, sep = "\t", quote = F) count <- pbmc[["RNA"]]@counts ## 原始的counts计数 count <- as.data.frame(count) mean <- apply(count, 1, mean) ## 按行求均值 variance <- apply(count, 1, var) ## 按行求方差 hvf.info <- data.frame(mean, variance) ## 生成数据框 hvf.info$variance.expected <- 0 ## 添加列并赋值 hvf.info$variance.standardized <- 0 not.const <- hvf.info$variance > 0 fit <- loess( ## 根据平均值和方差进行拟合 formula = log10(x = variance) ~ log10(x = mean), data = hvf.info[not.const, ], span = 0.3 ) hvf.info$variance.expected[not.const] <- 10 ^ fit$fitted ## 计算期望方差 count_std <- matrix(nrow = nrow(count), ncol = ncol(count)) ## 生成标准化计数数据框 identical(rownames(count), rownames(hvf.info)) clip_max <- sqrt(ncol(count_std)) ## 计算修剪的最大值 clip_max for (i in 1:nrow(count_std)) { count_std[i,] <- (as.numeric(count[i,]) - hvf.info[i,1])/sqrt(hvf.info[i,3]) ## 标准化计数 count_std[i,][count_std[i,] >=clip_max] <- clip_max ## 修剪数据 hvf.info[i,4] <- var(as.numeric(count_std[i,])) ## 计算标准化数据后的期望方差 } head(std) ## 流程中的标准结果 head(hvf.info) ## 手动生成的结果 verify <- round(std$vst.variance.standardized, 1) %in% round(hvf.info$variance.standardized, 1) table(verify) ## 统计验证结果,有一个不一致,可能是四舍五入问题
002、改善程序运行效率
dat <- read.table("dat.txt", row.names = 1, header = T) dim(dat) dat[1:5, 1:5] vst.mean <- apply(dat, 1, mean) vst.variance <- apply(dat, 1, var) hvf.info <- data.frame(vst.mean, vst.variance) hvf.info$variance.expected <- 0 hvf.info$variance.standardized <- 0 not.const <- hvf.info$vst.variance > 0 not.const head(hvf.info) fit <- loess( formula = log10(vst.variance) ~ log10(vst.mean), data = hvf.info[not.const, ], span = 0.3 ) hvf.info$variance.expected[not.const] <- 10 ^ fit$fitted head(hvf.info) max_chip <- sqrt(2638) head(hvf.info) a <- hvf.info[,1] b <- sqrt(hvf.info[,3]) dat <- t(dat) std_count <- dat for (i in 1:ncol(dat)) { std_count[,i] <- (dat[,i] - a[i])/b[i] std_count[,i][std_count[,i] >= max_chip] <- max_chip hvf.info[i, 4] <- var(std_count[,i]) } write.table(hvf.info, "xxx.txt", row.names = T, col.names = T, sep = "\t", quote = F)
参考:
001:https://www.jianshu.com/p/4f7aeae81ef1
002:https://zhuanlan.zhihu.com/p/479549742
003:https://doi.org/10.1016/j.cell.2019.05.031






【推荐】国内首个AI IDE,深度理解中文开发场景,立即下载体验Trae
【推荐】编程新体验,更懂你的AI,立即体验豆包MarsCode编程助手
【推荐】抖音旗下AI助手豆包,你的智能百科全书,全免费不限次数
【推荐】轻量又高性能的 SSH 工具 IShell:AI 加持,快人一步
· 震惊!C++程序真的从main开始吗?99%的程序员都答错了
· 【硬核科普】Trae如何「偷看」你的代码?零基础破解AI编程运行原理
· 单元测试从入门到精通
· 上周热点回顾(3.3-3.9)
· winform 绘制太阳,地球,月球 运作规律
2021-06-07 c语言 13-4
2021-06-07 c语言 13-3
2021-06-07 c语言 13-3
2021-06-07 c语言 13 - 3
2021-06-07 c语言中冒泡排序法