利用GEMMA进行GWAS分析
1、软件下载,下载地址:https://github.com/genetics-statistics/GEMMA/releases

下载过程:
root@PC1:/home/gemma_test# ls root@PC1:/home/gemma_test# pwd /home/gemma_test root@PC1:/home/gemma_test# wget https://github.com/genetics-statistics/GEMMA/releases/download/v0.98.5/gemma-0.98.5-linux-static-AMD64.gz ## 下载 --2021-12-25 22:02:46-- https://github.com/genetics-statistics/GEMMA/releases/download/v0.98.5/gemma-0.98.5-linux-static-AMD64.gz Resolving github.com (github.com)... 20.205.243.166 Connecting to github.com (github.com)|20.205.243.166|:443... connected. HTTP request sent, awaiting response... 302 Found Location: https://objects.githubusercontent.com/github-production-release-asset-2e65be/2880217/634e9bdc-e9f8-409d-8354-16874abd27ce?X-Amz-Algorithm=AWS4-HMAC-SHA256&X-Amz-Credential=AKIAIWNJYAX4CSVEH53A%2F20211225%2Fus-east-1%2Fs3%2Faws4_request&X-Amz-Date=20211225T140247Z&X-Amz-Expires=300&X-Amz-Signature=33e6f15d35852f87e55df57d93955d7ed3474f3185cae7fa05edbe0347b39d5d&X-Amz-SignedHeaders=host&actor_id=0&key_id=0&repo_id=2880217&response-content-disposition=attachment%3B%20filename%3Dgemma-0.98.5-linux-static-AMD64.gz&response-content-type=application%2Foctet-stream [following] --2021-12-25 22:02:47-- https://objects.githubusercontent.com/github-production-release-asset-2e65be/2880217/634e9bdc-e9f8-409d-8354-16874abd27ce?X-Amz-Algorithm=AWS4-HMAC-SHA256&X-Amz-Credential=AKIAIWNJYAX4CSVEH53A%2F20211225%2Fus-east-1%2Fs3%2Faws4_request&X-Amz-Date=20211225T140247Z&X-Amz-Expires=300&X-Amz-Signature=33e6f15d35852f87e55df57d93955d7ed3474f3185cae7fa05edbe0347b39d5d&X-Amz-SignedHeaders=host&actor_id=0&key_id=0&repo_id=2880217&response-content-disposition=attachment%3B%20filename%3Dgemma-0.98.5-linux-static-AMD64.gz&response-content-type=application%2Foctet-stream Resolving objects.githubusercontent.com (objects.githubusercontent.com)... 185.199.108.133, 185.199.111.133, 185.199.110.133, ... Connecting to objects.githubusercontent.com (objects.githubusercontent.com)|185.199.108.133|:443... connected. HTTP request sent, awaiting response... 200 OK Length: 6394866 (6.1M) [application/octet-stream] Saving to: ‘gemma-0.98.5-linux-static-AMD64.gz’ gemma-0.98.5-linux-static-AMD64.g 100%[==========================================================>] 6.10M 275KB/s in 22s 2021-12-25 22:03:10 (284 KB/s) - ‘gemma-0.98.5-linux-static-AMD64.gz’ saved [6394866/6394866] root@PC1:/home/gemma_test# ls gemma-0.98.5-linux-static-AMD64.gz root@PC1:/home/gemma_test# gunzip gemma-0.98.5-linux-static-AMD64.gz ## 解压 root@PC1:/home/gemma_test# ls gemma-0.98.5-linux-static-AMD64 root@PC1:/home/gemma_test# chmod 755 gemma-0.98.5-linux-static-AMD64 ## 赋予执行权限 root@PC1:/home/gemma_test# ls gemma-0.98.5-linux-static-AMD64
2、调用测试
root@PC1:/home/gemma_test# ls gemma-0.98.5-linux-static-AMD64 root@PC1:/home/gemma_test# ./gemma-0.98.5-linux-static-AMD64 GEMMA 0.98.5 (2021-08-25) by Xiang Zhou, Pjotr Prins and team (C) 2012-2021 type ./gemma -h [num] for detailed help options: 1: quick guide 2: file I/O related 3: SNP QC 4: calculate relatedness matrix 5: perform eigen decomposition 6: perform variance component estimation 7: fit a linear model 8: fit a linear mixed model 9: fit a multivariate linear mixed model 10: fit a Bayesian sparse linear mixed model 11: obtain predicted values 12: calculate snp variance covariance 13: note 14: debug options The GEMMA software is distributed under the GNU General Public v3 -license show license information see also http://www.xzlab.org/software.html, https://github.com/genetics-statistics
3、测试数据下载,地址:https://github.com/genetics-statistics/GEMMA/releases

root@PC1:/home/gemma_test# ls 2000.bed 2000.bim 2000.fam gemma-0.98.5-linux-static-AMD64
4、剔除没有表型记录的数据,gemma只分析有表型记录的数据
root@PC1:/home/gemma_test# ls 2000.bed 2000.bim 2000.fam gemma-0.98.5-linux-static-AMD64 root@PC1:/home/gemma_test# awk '$6 != "-9"' 2000.fam | awk '{print $1, $2}' > keep.txt ## 提取fam文件第六列不为-9的数据,然后提取前两列 root@PC1:/home/gemma_test# head keep.txt 88 88 108 108 139 139 159 159 265 265 350 350 351 351 403 403 410 410 424 424 root@PC1:/home/gemma_test# plink --bfile 2000 --keep keep.txt --make-bed --out 2000new ## 利用plink软件--keep命令提取没有缺失的数据 PLINK v1.90b6.24 64-bit (6 Jun 2021) www.cog-genomics.org/plink/1.9/ (C) 2005-2021 Shaun Purcell, Christopher Chang GNU General Public License v3 Logging to 2000new.log. Options in effect: --bfile 2000 --keep keep.txt --make-bed --out 2000new 15974 MB RAM detected; reserving 7987 MB for main workspace. 2000 variants loaded from .bim file. 1008 people (0 males, 0 females, 1008 ambiguous) loaded from .fam. Ambiguous sex IDs written to 2000new.nosex . 876 phenotype values loaded from .fam. --keep: 876 people remaining. Warning: Ignoring phenotypes of missing-sex samples. If you don't want those phenotypes to be ignored, use the --allow-no-sex flag. Using 1 thread (no multithreaded calculations invoked). Before main variant filters, 876 founders and 0 nonfounders present. Calculating allele frequencies... done. Total genotyping rate in remaining samples is 0.973096. 2000 variants and 876 people pass filters and QC. Phenotype data is quantitative. --make-bed to 2000new.bed + 2000new.bim + 2000new.fam ... done. root@PC1:/home/gemma_test# ls 2000.bed 2000.fam 2000new.bim 2000new.log gemma-0.98.5-linux-static-AMD64 2000.bim 2000new.bed 2000new.fam 2000new.nosex keep.txt
root@PC1:/home/gemma_test# wc -l 2000new.* 0 2000new.bed 2000 2000new.bim 876 2000new.fam 29 2000new.log 1008 2000new.nosex 3913 total
## 可见一共有2000个位点, 876个样本
5、质控
root@PC1:/home/gemma_test# ls 2000.bed 2000.fam 2000new.bim 2000new.log gemma-0.98.5-linux-static-AMD64 2000.bim 2000new.bed 2000new.fam 2000new.nosex keep.txt root@PC1:/home/gemma_test# plink --bfile 2000new --mind 0.1 --geno 0.05 --maf 0.01 --make-bed --out 2000new2 ## 样本缺失率大于10%提出,位点缺失率大于5%剔除,最小等位基因频率小于1%剔除
##gemma软件只会分析位点缺失率小于5%的位点,因此使用--geno 0.05的标准
PLINK v1.90b6.24 64-bit (6 Jun 2021) www.cog-genomics.org/plink/1.9/ (C) 2005-2021 Shaun Purcell, Christopher Chang GNU General Public License v3 Logging to 2000new2.log. Options in effect: --bfile 2000new --geno 0.05 --maf 0.01 --make-bed --mind 0.1 --out 2000new2 15974 MB RAM detected; reserving 7987 MB for main workspace. 2000 variants loaded from .bim file. 876 people (0 males, 0 females, 876 ambiguous) loaded from .fam. Ambiguous sex IDs written to 2000new2.nosex . 876 phenotype values loaded from .fam. Warning: Ignoring phenotypes of missing-sex samples. If you don't want those phenotypes to be ignored, use the --allow-no-sex flag. 36 people removed due to missing genotype data (--mind). IDs written to 2000new2.irem . Using 1 thread (no multithreaded calculations invoked). Before main variant filters, 840 founders and 0 nonfounders present. Calculating allele frequencies... done. Total genotyping rate in remaining samples is 0.978071. 15 variants removed due to missing genotype data (--geno). 0 variants removed due to minor allele threshold(s) (--maf/--max-maf/--mac/--max-mac). 1985 variants and 840 people pass filters and QC. Phenotype data is quantitative. --make-bed to 2000new2.bed + 2000new2.bim + 2000new2.fam ... done. root@PC1:/home/gemma_test# ls 2000.bed 2000.fam 2000new2.bim 2000new2.irem 2000new2.nosex 2000new.bim 2000new.log gemma-0.98.5-linux-static-AMD64 2000.bim 2000new2.bed 2000new2.fam 2000new2.log 2000new.bed 2000new.fam 2000new.nosex keep.txt
6、计算kinship矩阵
root@PC1:/home/gemma_test# ls 2000.bed 2000.fam 2000new2.bim 2000new2.irem 2000new2.nosex 2000new.bim 2000new.log gemma-0.98.5-linux-static-AMD64 2000.bim 2000new2.bed 2000new2.fam 2000new2.log 2000new.bed 2000new.fam 2000new.nosex keep.txt root@PC1:/home/gemma_test# ./gemma-0.98.5-linux-static-AMD64 -bfile 2000new2 -gk 2 -o kin ## -gk 2 表示生成的kinship进行标准化(scale),-o指定输出的名称 GEMMA 0.98.5 (2021-08-25) by Xiang Zhou, Pjotr Prins and team (C) 2012-2021 Reading Files ... ## number of total individuals = 840 ## number of analyzed individuals = 840 ## number of covariates = 1 ## number of phenotypes = 1 ## number of total SNPs/var = 1985 ## number of analyzed SNPs = 1985 Calculating Relatedness Matrix ... ================================================== 100% **** INFO: Done. root@PC1:/home/gemma_test# ls 2000.bed 2000new2.bed 2000new2.irem 2000new.bed 2000new.log keep.txt 2000.bim 2000new2.bim 2000new2.log 2000new.bim 2000new.nosex output 2000.fam 2000new2.fam 2000new2.nosex 2000new.fam gemma-0.98.5-linux-static-AMD64 root@PC1:/home/gemma_test# ls output/ kin.log.txt kin.sXX.txt
7、将kinship文件移动至当前目录
root@PC1:/home/gemma_test# ls 2000.bed 2000new2.bed 2000new2.irem 2000new.bed 2000new.log keep.txt 2000.bim 2000new2.bim 2000new2.log 2000new.bim 2000new.nosex output 2000.fam 2000new2.fam 2000new2.nosex 2000new.fam gemma-0.98.5-linux-static-AMD64 root@PC1:/home/gemma_test# mv ./output/kin.sXX.txt . root@PC1:/home/gemma_test# ls 2000.bed 2000new2.bed 2000new2.irem 2000new.bed 2000new.log keep.txt 2000.bim 2000new2.bim 2000new2.log 2000new.bim 2000new.nosex kin.sXX.txt 2000.fam 2000new2.fam 2000new2.nosex 2000new.fam gemma-0.98.5-linux-static-AMD64 output
8、进行GWAS分析
root@PC1:/home/gemma_test# ls 2000.bed 2000new2.bed 2000new2.irem 2000new.bed 2000new.log keep.txt 2000.bim 2000new2.bim 2000new2.log 2000new.bim 2000new.nosex kin.sXX.txt 2000.fam 2000new2.fam 2000new2.nosex 2000new.fam gemma-0.98.5-linux-static-AMD64 output root@PC1:/home/gemma_test# ./gemma-0.98.5-linux-static-AMD64 -bfile 2000new2 -k kin.sXX.txt -lmm 1 -o result ## -k 指定kinship文件,
##-lmm 进行混合线性模型GWAS分析。选项代表不同的检验方式:1为Wald检验;2为似然比检验;3为score检验;4为以上三个检验全部执行。一般使用wald检验即可 GEMMA 0.98.5 (2021-08-25) by Xiang Zhou, Pjotr Prins and team (C) 2012-2021 Reading Files ... ## number of total individuals = 840 ## number of analyzed individuals = 840 ## number of covariates = 1 ## number of phenotypes = 1 ## number of total SNPs/var = 1985 ## number of analyzed SNPs = 1985 Start Eigen-Decomposition... **** WARNING: Matrix G has 3 eigenvalues close to zero pve estimate =0.593312 se(pve) =0.0603374 ================================================== 100% **** INFO: Done. root@PC1:/home/gemma_test# ls 2000.bed 2000new2.bed 2000new2.irem 2000new.bed 2000new.log keep.txt 2000.bim 2000new2.bim 2000new2.log 2000new.bim 2000new.nosex kin.sXX.txt 2000.fam 2000new2.fam 2000new2.nosex 2000new.fam gemma-0.98.5-linux-static-AMD64 output root@PC1:/home/gemma_test# ls output/ kin.log.txt result.assoc.txt result.log.txt root@PC1:/home/gemma_test# head output/result.assoc.txt ## 查看结果文件的前10行,重要信息在 chr、rs、ps、p_wald列 chr rs ps n_miss allele1 allele0 af beta se logl_H1 l_remle p_wald 1 32 363 42 G C 0.065 -3.817377e+00 2.334342e+00 -3.477735e+03 1.462921e+00 1.023592e-01 1 118 1645 38 T G 0.175 -1.646150e+00 1.631679e+00 -3.478488e+03 1.445743e+00 3.133288e-01 1 119 1646 34 G A 0.171 -1.546425e+00 1.614092e+00 -3.478554e+03 1.449508e+00 3.383002e-01 1 134 2309 17 A T 0.281 1.132549e+00 1.905078e+00 -3.478484e+03 1.448278e+00 5.523452e-01 1 135 2310 21 A C 0.282 1.608435e-01 1.908279e+00 -3.478656e+03 1.470582e+00 9.328482e-01 1 144 2554 25 T C 0.058 -1.837053e+00 2.161237e+00 -3.478834e+03 1.461591e+00 3.955674e-01 1 148 2673 17 T C 0.083 -2.581828e+00 1.941161e+00 -3.478247e+03 1.451538e+00 1.838659e-01 1 162 3102 27 G A 0.311 3.694870e+00 1.470007e+00 -3.475745e+03 1.456267e+00 1.214033e-02 1 182 4648 14 A C 0.297 3.429882e+00 1.930098e+00 -3.477053e+03 1.436600e+00 7.592262e-02
9、使用R绘制曼哈顿图
##install.packages("qqman") library(qqman) ## 加载qqman包 results_log <- read.table("result.assoc.txt", head=TRUE) ## 读取结果文件 manhattan(results_log,chr="chr",bp="ps",p="p_wald",snp="rs", ylim = c(0, 10), cex = 2, cex.axis = 1.5, col = 1, suggestiveline = F, genomewideline = F, main = "Manhattan plot") ## 绘制曼哈顿图

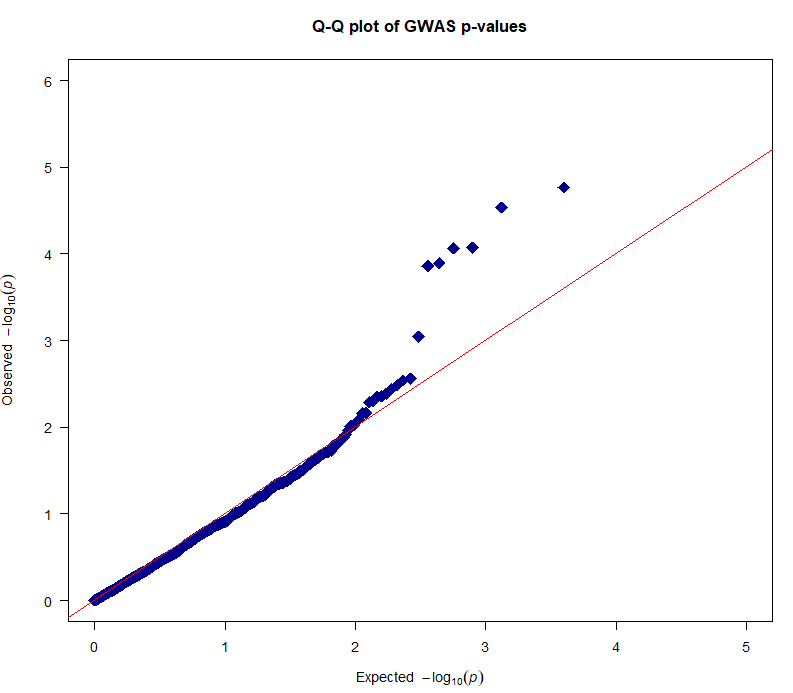
10、绘制qq图
qq(results_log$p_wald, main = "Q-Q plot of GWAS p-values", xlim = c(0, 5), ylim = c(0, 6), pch = 18, col = "blue4", cex = 2, las = 1)
参考:
https://www.jianshu.com/p/d31404620c9b
https://www.jianshu.com/p/372ec8585b49
分类:
生信






【推荐】国内首个AI IDE,深度理解中文开发场景,立即下载体验Trae
【推荐】编程新体验,更懂你的AI,立即体验豆包MarsCode编程助手
【推荐】抖音旗下AI助手豆包,你的智能百科全书,全免费不限次数
【推荐】轻量又高性能的 SSH 工具 IShell:AI 加持,快人一步
· 基于Microsoft.Extensions.AI核心库实现RAG应用
· Linux系列:如何用heaptrack跟踪.NET程序的非托管内存泄露
· 开发者必知的日志记录最佳实践
· SQL Server 2025 AI相关能力初探
· Linux系列:如何用 C#调用 C方法造成内存泄露
· 震惊!C++程序真的从main开始吗?99%的程序员都答错了
· 【硬核科普】Trae如何「偷看」你的代码?零基础破解AI编程运行原理
· 单元测试从入门到精通
· 上周热点回顾(3.3-3.9)
· winform 绘制太阳,地球,月球 运作规律
2020-12-25 C语言获取数值的最后几位数