Topic extraction with Non-negative Matrix Factorization and Latent Dirichlet Allocation of sklearn
Topic extraction with Non-negative Matrix Factorization and Latent Dirichlet Allocation
https://scikit-learn.org/stable/auto_examples/applications/plot_topics_extraction_with_nmf_lda.html#sphx-glr-auto-examples-applications-plot-topics-extraction-with-nmf-lda-py
使用非负矩阵分解方法, 对于文档库进行分析, 提取语料库话题结构的累加模型。
输出绘制的图像是关于 各个话题, 每个话题绘制最高的介个单词, 基于矩阵分解后的话题特征系数。
This is an example of applying
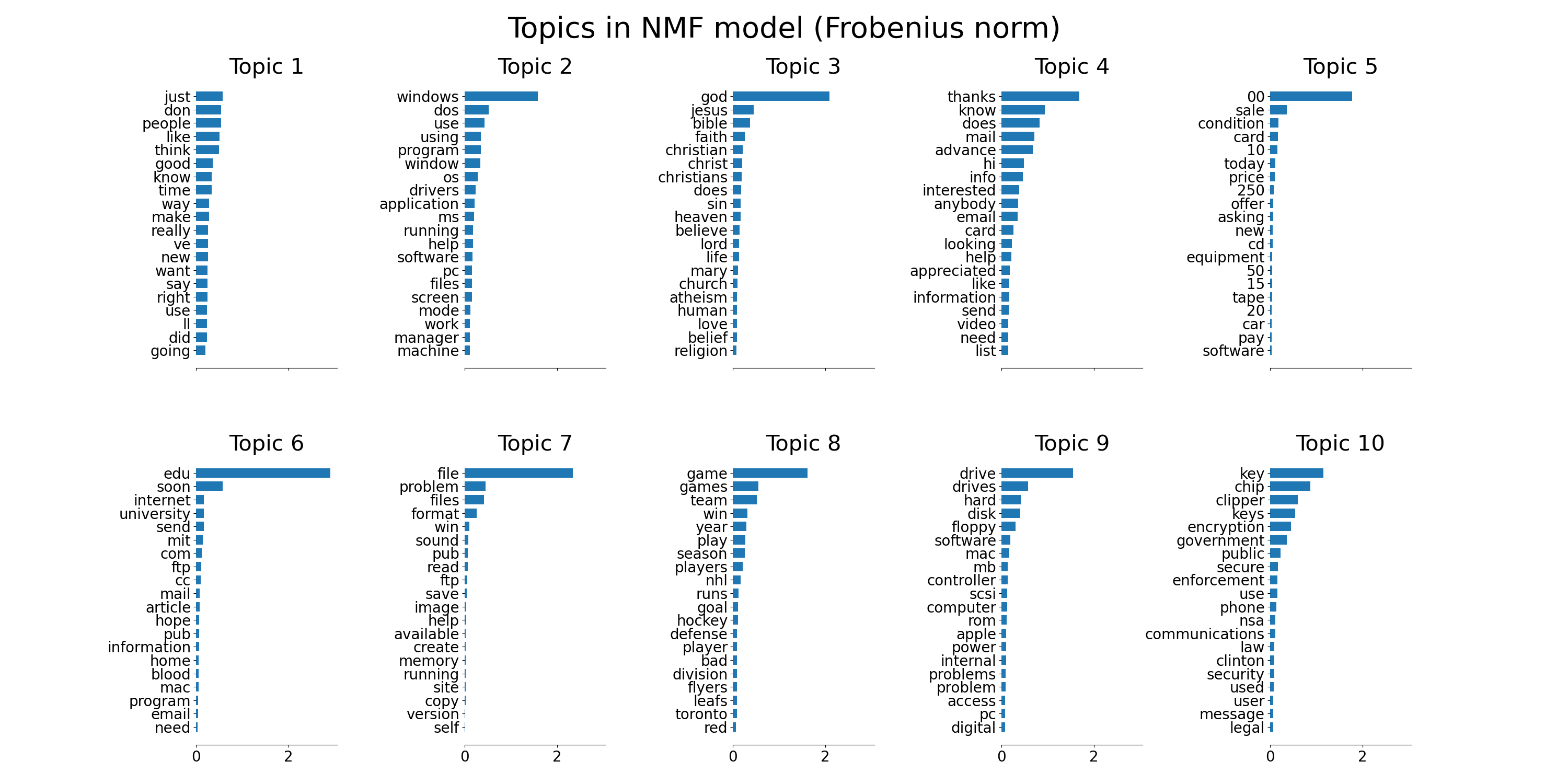
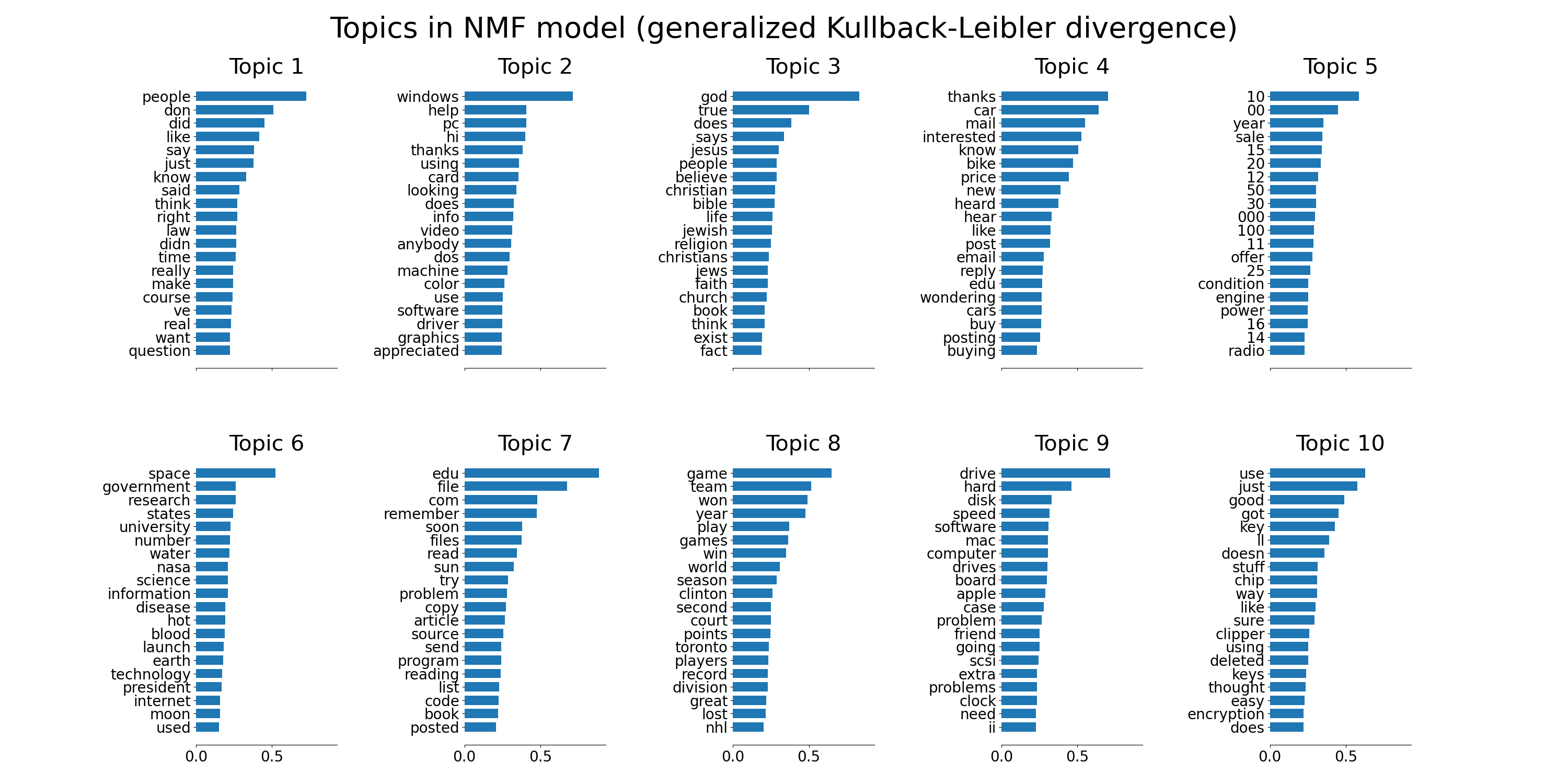
NMFandLatentDirichletAllocationon a corpus of documents and extract additive models of the topic structure of the corpus. The output is a plot of topics, each represented as bar plot using top few words based on weights.Non-negative Matrix Factorization is applied with two different objective functions: the Frobenius norm, and the generalized Kullback-Leibler divergence. The latter is equivalent to Probabilistic Latent Semantic Indexing.
The default parameters (n_samples / n_features / n_components) should make the example runnable in a couple of tens of seconds. You can try to increase the dimensions of the problem, but be aware that the time complexity is polynomial in NMF. In LDA, the time complexity is proportional to (n_samples * iterations).
Code
"""
=======================================================================================
Topic extraction with Non-negative Matrix Factorization and Latent Dirichlet Allocation
=======================================================================================
This is an example of applying :class:`~sklearn.decomposition.NMF` and
:class:`~sklearn.decomposition.LatentDirichletAllocation` on a corpus
of documents and extract additive models of the topic structure of the
corpus. The output is a plot of topics, each represented as bar plot
using top few words based on weights.
Non-negative Matrix Factorization is applied with two different objective
functions: the Frobenius norm, and the generalized Kullback-Leibler divergence.
The latter is equivalent to Probabilistic Latent Semantic Indexing.
The default parameters (n_samples / n_features / n_components) should make
the example runnable in a couple of tens of seconds. You can try to
increase the dimensions of the problem, but be aware that the time
complexity is polynomial in NMF. In LDA, the time complexity is
proportional to (n_samples * iterations).
"""
# Author: Olivier Grisel <olivier.grisel@ensta.org>
# Lars Buitinck
# Chyi-Kwei Yau <chyikwei.yau@gmail.com>
# License: BSD 3 clause
from time import time
import matplotlib.pyplot as plt
from sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer
from sklearn.decomposition import NMF, LatentDirichletAllocation
from sklearn.datasets import fetch_20newsgroups
n_samples = 2000
n_features = 1000
n_components = 10
n_top_words = 20
def plot_top_words(model, feature_names, n_top_words, title):
fig, axes = plt.subplots(2, 5, figsize=(30, 15), sharex=True)
axes = axes.flatten()
for topic_idx, topic in enumerate(model.components_):
print("------ topic ----------")
print(topic)
print("------ topic.argsort() ----------")
# get sorted index array from small to large
print(topic.argsort())
# get n_top_words words from tail
top_features_ind = topic.argsort()[:-n_top_words - 1:-1]
print("----- top_features_ind ---------")
print(top_features_ind)
top_features = [feature_names[i] for i in top_features_ind]
print("-------- top features -----------")
print(top_features)
weights = topic[top_features_ind]
print("----------- weights -------------")
print(weights)
ax = axes[topic_idx]
ax.barh(top_features, weights, height=0.7)
ax.set_title(f'Topic {topic_idx +1}',
fontdict={'fontsize': 30})
ax.invert_yaxis()
ax.tick_params(axis='both', which='major', labelsize=20)
for i in 'top right left'.split():
ax.spines[i].set_visible(False)
fig.suptitle(title, fontsize=40)
plt.subplots_adjust(top=0.90, bottom=0.05, wspace=0.90, hspace=0.3)
plt.show()
# Load the 20 newsgroups dataset and vectorize it. We use a few heuristics
# to filter out useless terms early on: the posts are stripped of headers,
# footers and quoted replies, and common English words, words occurring in
# only one document or in at least 95% of the documents are removed.
print("Loading dataset...")
t0 = time()
data, _ = fetch_20newsgroups(shuffle=True, random_state=1,
remove=('headers', 'footers', 'quotes'),
return_X_y=True)
data_samples = data[:n_samples]
print("done in %0.3fs." % (time() - t0))
# Use tf-idf features for NMF.
print("Extracting tf-idf features for NMF...")
tfidf_vectorizer = TfidfVectorizer(max_df=0.95, min_df=2,
max_features=n_features,
stop_words='english')
t0 = time()
tfidf = tfidf_vectorizer.fit_transform(data_samples)
print("done in %0.3fs." % (time() - t0))
# Use tf (raw term count) features for LDA.
print("Extracting tf features for LDA...")
tf_vectorizer = CountVectorizer(max_df=0.95, min_df=2,
max_features=n_features,
stop_words='english')
t0 = time()
tf = tf_vectorizer.fit_transform(data_samples)
print("done in %0.3fs." % (time() - t0))
print()
# Fit the NMF model
print("Fitting the NMF model (Frobenius norm) with tf-idf features, "
"n_samples=%d and n_features=%d..."
% (n_samples, n_features))
t0 = time()
nmf = NMF(n_components=n_components, random_state=1,
alpha=.1, l1_ratio=.5).fit(tfidf)
print("done in %0.3fs." % (time() - t0))
tfidf_feature_names = tfidf_vectorizer.get_feature_names()
plot_top_words(nmf, tfidf_feature_names, n_top_words,
'Topics in NMF model (Frobenius norm)')
# Fit the NMF model
print('\n' * 2, "Fitting the NMF model (generalized Kullback-Leibler "
"divergence) with tf-idf features, n_samples=%d and n_features=%d..."
% (n_samples, n_features))
t0 = time()
nmf = NMF(n_components=n_components, random_state=1,
beta_loss='kullback-leibler', solver='mu', max_iter=1000, alpha=.1,
l1_ratio=.5).fit(tfidf)
print("done in %0.3fs." % (time() - t0))
tfidf_feature_names = tfidf_vectorizer.get_feature_names()
plot_top_words(nmf, tfidf_feature_names, n_top_words,
'Topics in NMF model (generalized Kullback-Leibler divergence)')
print('\n' * 2, "Fitting LDA models with tf features, "
"n_samples=%d and n_features=%d..."
% (n_samples, n_features))
lda = LatentDirichletAllocation(n_components=n_components, max_iter=5,
learning_method='online',
learning_offset=50.,
random_state=0)
t0 = time()
lda.fit(tf)
print("done in %0.3fs." % (time() - t0))
tf_feature_names = tf_vectorizer.get_feature_names()
plot_top_words(lda, tf_feature_names, n_top_words, 'Topics in LDA model')
Output
Loading dataset...
done in 1.143s.
Extracting tf-idf features for NMF...
done in 0.285s.
Extracting tf features for LDA...
done in 0.278s.
Fitting the NMF model (Frobenius norm) with tf-idf features, n_samples=2000 and n_features=1000...
/home/circleci/project/sklearn/decomposition/_nmf.py:312: FutureWarning: The 'init' value, when 'init=None' and n_components is less than n_samples and n_features, will be changed from 'nndsvd' to 'nndsvda' in 1.1 (renaming of 0.26).
warnings.warn(("The 'init' value, when 'init=None' and "
done in 0.238s.
Fitting the NMF model (generalized Kullback-Leibler divergence) with tf-idf features, n_samples=2000 and n_features=1000...
/home/circleci/project/sklearn/decomposition/_nmf.py:312: FutureWarning: The 'init' value, when 'init=None' and n_components is less than n_samples and n_features, will be changed from 'nndsvd' to 'nndsvda' in 1.1 (renaming of 0.26).
warnings.warn(("The 'init' value, when 'init=None' and "
done in 0.816s.
Fitting LDA models with tf features, n_samples=2000 and n_features=1000...
done in 3.541s.
NMF of sklearn
https://scikit-learn.org/stable/modules/generated/sklearn.decomposition.NMF.html#sklearn.decomposition.NMF
对于非负矩阵X, 找到两个同样的非负矩阵, W 和 H, 使得 W和H的乘积 是 X的逼近值。
此工具被用于 维度约减, 源分解,话题提取。
Non-Negative Matrix Factorization (NMF).
Find two non-negative matrices (W, H) whose product approximates the non- negative matrix X. This factorization can be used for example for dimensionality reduction, source separation or topic extraction.
>>> import numpy as np >>> X = np.array([[1, 1], [2, 1], [3, 1.2], [4, 1], [5, 0.8], [6, 1]]) >>> from sklearn.decomposition import NMF >>> model = NMF(n_components=2, init='random', random_state=0) >>> W = model.fit_transform(X) >>> H = model.components_
Non-negative matrix factorization
https://en.wikipedia.org/wiki/Non-negative_matrix_factorization
NMF是对多道变量分析的一组算法,是一种逼近的分解算法, 非精确分解。
应用于 天文 计算机视觉 文档聚类 缺失值填补 化学测量 声音信号处理 推荐系统 生物信息学。
Non-negative matrix factorization (NMF or NNMF), also non-negative matrix approximation[1][2] is a group of algorithms in multivariate analysis and linear algebra where a matrix V is factorized into (usually) two matrices W and H, with the property that all three matrices have no negative elements. This non-negativity makes the resulting matrices easier to inspect. Also, in applications such as processing of audio spectrograms or muscular activity, non-negativity is inherent to the data being considered. Since the problem is not exactly solvable in general, it is commonly approximated numerically.
NMF finds applications in such fields as astronomy,[3][4] computer vision, document clustering,[1] missing data imputation,[5] chemometrics, audio signal processing, recommender systems,[6][7] and bioinformatics.[8
Illustration of approximate non-negative matrix factorization: the matrix V is represented by the two smaller matrices W and H, which, when multiplied, approximately reconstruct V.
https://blog.acolyer.org/2019/02/18/the-why-and-how-of-nonnegative-matrix-factorization/
文本挖掘使用词袋矩阵表示方法, 使用NMF分解。
W[R * T] 表示词袋特征空间 中, 关于特征的 权值矩阵, 其中每一行表示一个话题, 话题对于所有词都有一个权值 , W(r, t) --- 表示第r个话题关于第t个词的权重系数。R -- 共R个话题, T 表示共T个词。
H[D*R] 表示在词袋的特征空间中, 每一个文档对应的偏置值(bias), H(d, r) --- 表示第d个文档关于第r个话题的偏置值。
D-- 共D个文档, R -- 共R个话题。
In text mining consider the bag-of-words matrix representation where each row corresponds to a word, and each column to a document (for the attentive reader, that’s the transpose of the bag-of-words matrix we looked at in ‘Beyond news content’9).
NMF will produce two matrices W and H. The columns of W can be interpreted as basis documents (bags of words). What interpretation can we give to such a basis document in this case? They represent topics! Sets of words found simultaneously in different documents. H tells us how to sum contributions from different topics to reconstruct the word mix of a given original document.
Therefore, given a set of documents, NMF identifies topics and simultaneously classifies the documents among these different topics.
numpy usage
argsort
https://numpy.org/doc/stable/reference/generated/numpy.argsort.html
只返回原始数组值按照升序排序的 下表数组, 对原始数组不排序。
Returns the indices that would sort an array.
Perform an indirect sort along the given axis using the algorithm specified by the kind keyword. It returns an array of indices of the same shape as a that index data along the given axis in sorted order.
x = np.array([3, 1, 2])
np.argsort(x)
array([1, 2, 0])
Negative Indexing
https://numpy.org/doc/stable/reference/arrays.indexing.html
对于索引结构中第三个数, step, 如果是正的, 则向后跨步, 如果是负值, 则向前跨步。
The basic slice syntax is
i:j:kwhere i is the starting index, j is the stopping index, and k is the step (). This selects the m elements (in the corresponding dimension) with index values i, i + k, …, i + (m - 1) k where
and q and r are the quotient and remainder obtained by dividing j - i by k: j - i = q k + r, so that i + (m - 1) k < j.
Negative i and j are interpreted as n + i and n + j where n is the number of elements in the corresponding dimension. Negative k makes stepping go towards smaller indices.
Example
x = np.array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9])
x[1:7:2]
array([1, 3, 5])
x[-2:10] array([8, 9]) x[-3:3:-1] array([7, 6, 5, 4])




 浙公网安备 33010602011771号
浙公网安备 33010602011771号