24-有参转录组实战10-差异基因KEGG富集分析

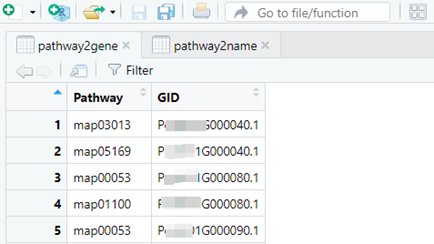
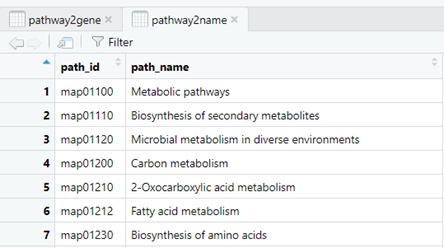
##########下面做KEGG######## emapper <- read.delim("out.emapper.annotations") emapper[emapper=="-"] <- NA#change "-" to "NA" emapper <- emapper[-(49584:49586),]#remove the final 3 rows DE <- read.delim("DE_genes_filter.txt") pathway2gene <- dplyr::select(emapper,Pathway=KEGG_Pathway, GID=query)%>% separate_rows(Pathway, sep = ',', convert = F)%>% filter(str_detect(Pathway, 'ko'))%>% mutate(Pathway= str_remove(Pathway, 'ko')) pathway2gene <- pathway2gene%>%dplyr::mutate(Pathway=paste0("map",Pathway))#前面加"map" library(magrittr) get_path2name <- function(){ keggpathid2name.df <- clusterProfiler:::kegg_list("pathway") keggpathid2name.df[,1] %>% gsub("path:map", "", .) colnames(keggpathid2name.df) <- c("path_id","path_name") return(keggpathid2name.df) } pathway2name <- get_path2name()
#查看下变量pathway2gene和pathway2name
ekp <- enricher(gene = DE$GID, TERM2GENE = pathway2gene, TERM2NAME = pathway2name, pvalueCutoff = 0.05, qvalueCutoff = 0.05) write.table(ekp, file = "Ptri_KEGG_test",sep = '\t',quote = F) #这里富集出来了,通路很少,可以自己调整P值和Q值的参数,或者在差异基因的筛选条件上放宽一点。Dsecription是对应https://rest.kegg.jp/list/pathway/#另外GeneRatio做成百分比
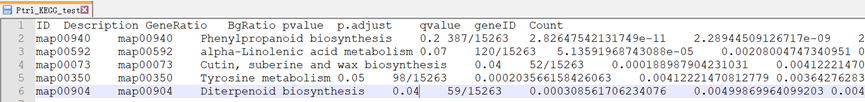
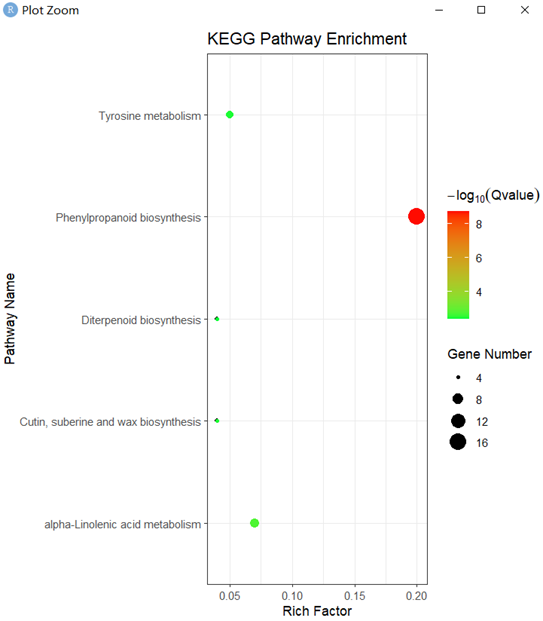
ekp2 <- read.delim("Ptri_KEGG_test") library(ggplot2) pp <- ggplot(ekp2, aes(GeneRatio, Description)) pp + geom_point() + geom_point(aes(size = Count)) + geom_point(aes(size = Count, color = -1 * log10(qvalue))) + scale_colour_gradient(low = "green", high = "red") + labs(color = expression(-log[10](Qvalue)), size = "Gene Number", x = "Rich Factor", y = "Pathway Name", title = "KEGG Pathway Enrichment") + theme_bw() #图片自己慢慢修,更多图表自行百度,可参考教程https://zhuanlan.zhihu.com/p/377356510
#加个尾图

#魔法少女小圆~


 KEGG富集分析
KEGG富集分析



【推荐】国内首个AI IDE,深度理解中文开发场景,立即下载体验Trae
【推荐】编程新体验,更懂你的AI,立即体验豆包MarsCode编程助手
【推荐】抖音旗下AI助手豆包,你的智能百科全书,全免费不限次数
【推荐】轻量又高性能的 SSH 工具 IShell:AI 加持,快人一步
· 震惊!C++程序真的从main开始吗?99%的程序员都答错了
· winform 绘制太阳,地球,月球 运作规律
· 【硬核科普】Trae如何「偷看」你的代码?零基础破解AI编程运行原理
· 上周热点回顾(3.3-3.9)
· 超详细:普通电脑也行Windows部署deepseek R1训练数据并当服务器共享给他人