Cistrome数据库挖掘转录因子靶基因 | TF | target
进入http://cistrome.org/db/#/数据库
输入你感兴趣的转录因子,比如HDAC1
选择物种,以及你感兴趣的cell type
点击可以查询”check a putative target“
也可以下载全部的”Putative Target“
最后个性化绘图
总结:
适合辅助分析,提供额外的fancy的证据,还是有一定的power的。
绘图代码:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 | library(ggplot2)library(RColorBrewer)Hdac1.chipseq.1 <- read.csv("ChIPseq/Hdac1_targets_mm_ESC_CR.txt", sep = "\t", stringsAsFactors = F)Hdac1.chipseq.1$humanGene <- toupper(Hdac1.chipseq.1$symbol)Hdac1.chipseq.1$cellLine <- "mm_ESC_CR"Hdac1.chipseq.1$CellType <- "Embryonic Stem Cell"Hdac1.chipseq <- rbind(Hdac1.chipseq.1, Hdac1.chipseq.2, Hdac1.chipseq.3, Hdac1.chipseq.4, Hdac1.chipseq.5)dim(Hdac1.chipseq)# sort scoreHdac1.chipseq <- Hdac1.chipseq[order(Hdac1.chipseq$score, decreasing = T),]# remove duplicatesHdac1.chipseq <- Hdac1.chipseq[!duplicated(Hdac1.chipseq[,c("chrom","symbol","cellLine")]),]core.risk.genes <- c("GLI3","BCL11A","FOXO1","HEY1","HEY2","SOX11","ASCL1","TUBB3","TFAP2A","NR2F2")Hdac1.chipseq.core <- subset(Hdac1.chipseq, humanGene %in% core.risk.genes & score > 0)Hdac1.chipseq.core$humanGene <- factor(Hdac1.chipseq.core$humanGene, levels = unique(Hdac1.chipseq.core$humanGene))Hdac1.chipseq.core$CellType <- factor(Hdac1.chipseq.core$CellType, levels = c("iPSC", "Embryonic Stem Cell", "Embryonic Fibroblast Cell", "Epithelium"))options(repr.plot.width=7, repr.plot.height=5)p <- ggplot(Hdac1.chipseq.core, aes(x=humanGene, y=score, color=CellType)) + # facet_wrap(~variable, ncol = 3) + theme_bw() + # geom_violin(trim=T, scale="width") + # aes(fill=group) # width, area, count # geom_boxplot() + geom_jitter(position=position_jitter(0),aes(fill=CellType),shape = 21,colour = "grey50",size = 4,stroke = 1) + labs(title="HDAC1",x="", y = "Score from ChIP-seq") + # theme(legend.title=element_blank()) + # just remove inside grid theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank()) + # remove top and right border theme(axis.line = element_line(colour = "black"), panel.grid.major = element_blank(), panel.grid.minor = element_blank(), panel.border = element_blank(), panel.background = element_blank()) + # force y start from 0 # scale_y_continuous(expand = c(0, 0), limits = c(-0.3, 11.5)) + # title position theme(plot.title = element_text(hjust = 0.5, face = "italic", size=20)) + ## theme(# legend.position = "none", axis.text.x = element_text(angle = 60, size = 12, vjust = 0.5, face = "italic"), axis.title.y = element_text(size = 16)) + scale_color_manual(values=brewer.pal(8,"Set1")) + scale_fill_manual(values=brewer.pal(8,"Set1"))pggsave(filename = "../manuscript/HDAC1.target.score.pdf", width = 7, height = 5) |
成品图
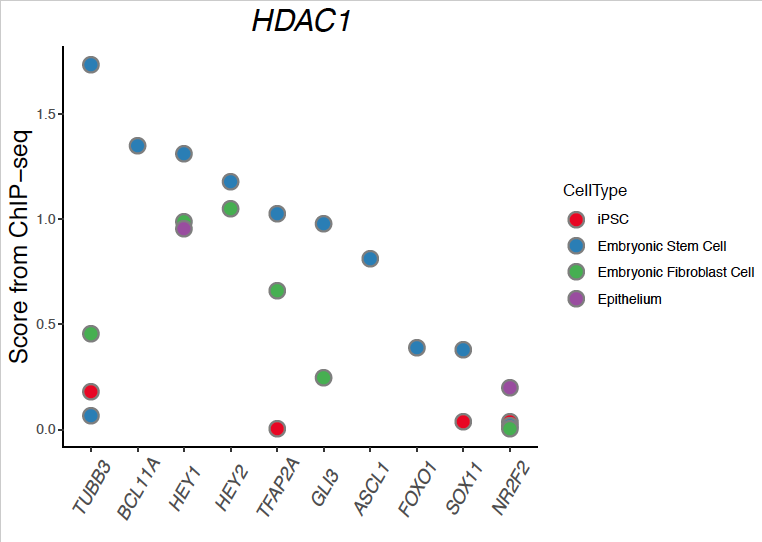
看着还不错吧,可以扯上一扯。
参考目录:human/singleCell/HSCR/HDAC/HDAC.ipynb
标签:
转录调控


【推荐】国内首个AI IDE,深度理解中文开发场景,立即下载体验Trae
【推荐】编程新体验,更懂你的AI,立即体验豆包MarsCode编程助手
【推荐】抖音旗下AI助手豆包,你的智能百科全书,全免费不限次数
【推荐】轻量又高性能的 SSH 工具 IShell:AI 加持,快人一步
· 从 HTTP 原因短语缺失研究 HTTP/2 和 HTTP/3 的设计差异
· AI与.NET技术实操系列:向量存储与相似性搜索在 .NET 中的实现
· 基于Microsoft.Extensions.AI核心库实现RAG应用
· Linux系列:如何用heaptrack跟踪.NET程序的非托管内存泄露
· 开发者必知的日志记录最佳实践
· winform 绘制太阳,地球,月球 运作规律
· AI与.NET技术实操系列(五):向量存储与相似性搜索在 .NET 中的实现
· 超详细:普通电脑也行Windows部署deepseek R1训练数据并当服务器共享给他人
· 【硬核科普】Trae如何「偷看」你的代码?零基础破解AI编程运行原理
· 上周热点回顾(3.3-3.9)
2019-03-31 GT sport赛道详解 - Dragon Trail | 龙之径
2017-03-31 CNV