HDAC | EZH2 | 常见基因组功能数据介绍 | 表观注释 | DNase-seq | ChIP-Seq | ATAC-Seq | eQTL | ENCODE | ROADMAP
一图胜千言
Characterizing cis-regulatory elements using single-cell epigenomics - 2022
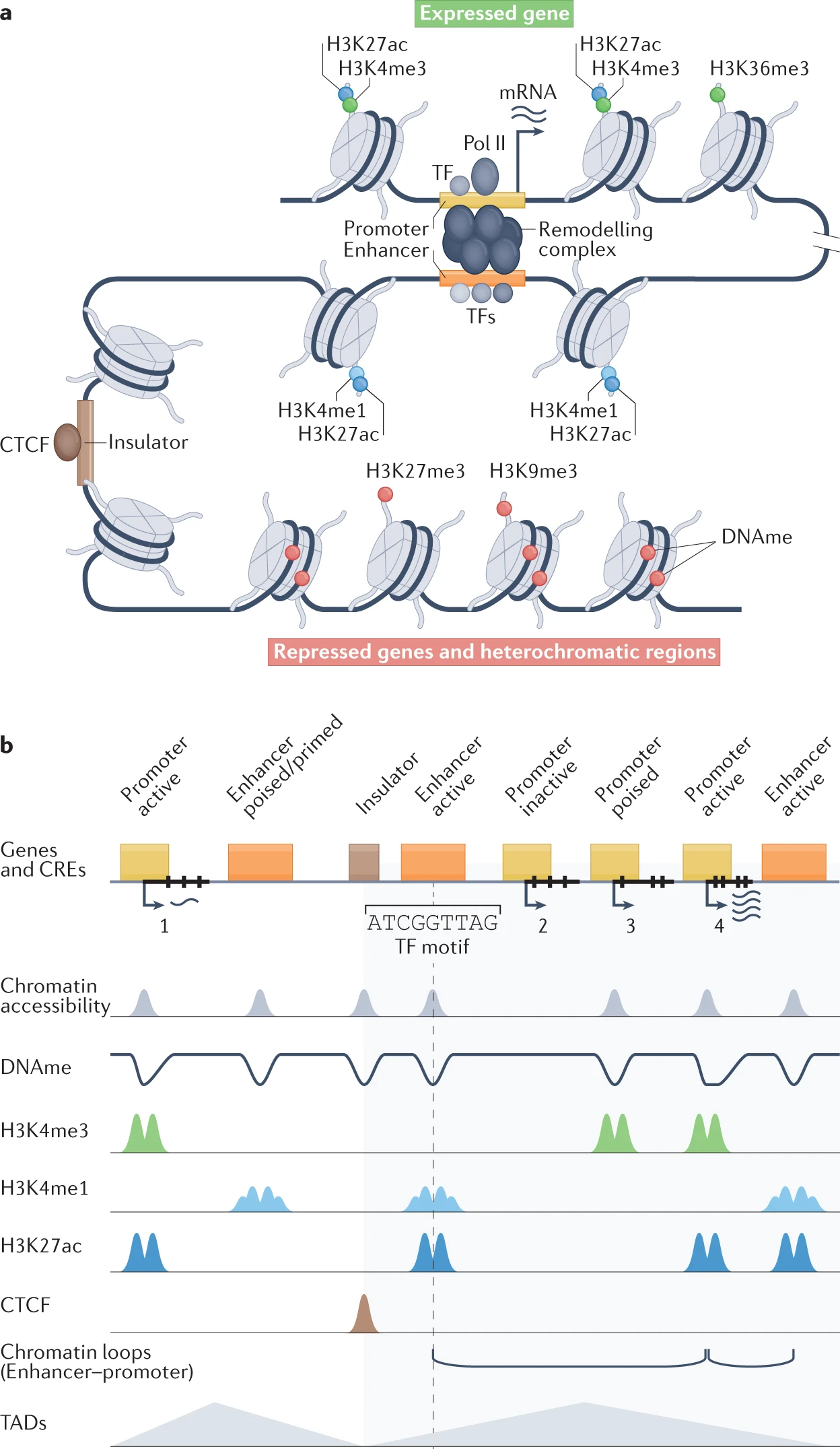
2022-06-15
最近在写HDAC1和EZH2的discussion,需要深入到表观数据的分析。
DC就是deacetylation of histone tails,那你知道具体DC哪些地方吗?那就得了解nucleosome和histone的构造,
H3指的就是哪一个histone,K或者就是指histone tail上的氨基酸,最后就是修饰的类型,me可能会有三次,所以有me3,对这种命名体系一定要了熟于心。
nucleosome构造:只有四种histone,H2A, H2B, H3 and H4,Each nucleosome consists of two subunits, both made of histones H2A, H2B, H3 and H4,相当于重复了一次。

histone的构造
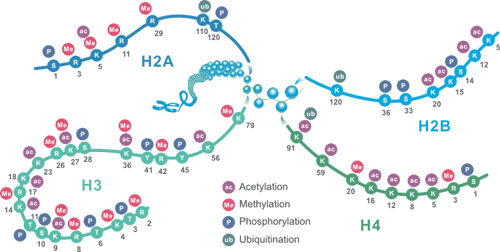
参考:
- Four Common Histone Modifications
- H3K27 acetylation and gene expression analysis reveals differences in placental chromatin activity in fetal growth restriction
- Chemical genomics reveals histone deacetylases are required for core regulatory transcription
- Histone deacetylase (HDAC) 1 and 2 are essential for accurate cell division and the pluripotency of embryonic stem cells
functional genomics | epigenomic annotations
刚入行的总是一头雾水,对这些表观的标记一点兴趣都没有,种类繁多,总是记不住,这里我就做一个常识性的总结,不搞太多术语。
需要了解的也不多,常见的就那么几个,搞懂ENCODE和ROADMAP上有的就行。细节颇多,需要一点耐心。
各种类型的数据可以直接在这个genome browser里浏览:http://genomebrowser.wustl.edu/
注意:
- 所有的表观或转录组都具有非常强的组织(cell type)特异性
- ChIP-seq最大的特点就是需要input,作为对照
- ChIP-seq可以Identify direct and indirect protein-DNA interactions
- ChIP-seq preferred for functional information
原始数据:
- DHSs
- H3K4me3
- H3K9ac
- H3K27ac
- H3K4me1
处理后数据:
- Enhancer
- TFBSs
主要是ChIP-seq(immunological assays)占了很大一类,把它搞懂就行。
另一类non-immunological assays:ATAC-seq, MNase-seq, DNase-seq, and FAIRE-seq。
DHSs
DNase I hypersensitive site
DNase-seq
FAIRE-Seq is a successor
genome-wide DNA footprints
Deoxyribonuclease 脱氧核糖核酸酶
DNase I hypersensitive sites (DHSs) are regions of chromatin that are sensitive to cleavage by the DNase I enzyme. In these specific regions of the genome, chromatin has lost its condensed structure, exposing the DNA and making it accessible. This raises the availability of DNA to degradation by enzymes, such as DNase I. These accessible chromatin zones are functionally related to transcriptional activity, since this remodeled state is necessary for the binding of proteins such as transcription factors.

ChIP-seq
Basically,
- "encc-enhancer.bed" is enhancers defined with H3K27ac & H3K4me1 activity
- "encc-enhancer-atac.bed" is enhancers defined with H3K27ac & H3K4me1 activity as well as open chromatin (ATAC-seq) signal summits.
不同ChIP-seq的功能,一图胜千言:【我们用了第一行和最后一行,效率最高】
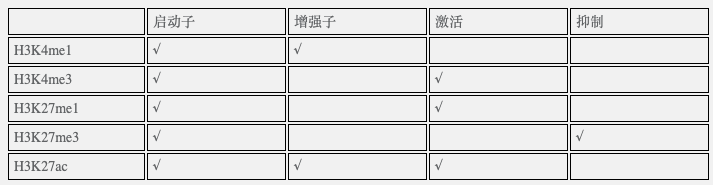
不同表观注释的比较:

待续~
快速使用epigenomic annotations data:
有个叫做baseline_v1.1的文件,里面包含了各种整理好的表观注释数据。
1 | https://data.broadinstitute.org/alkesgroup/LDSCORE/baseline_v1.1_bedfiles.tgz |
1 | ~/project2/CPloci/Evo/ENCODE/ |
包含的数据类型:
- Coding
- Intron
- Transcribe
- Conserved
- DGF
- DHS
- H3K9ac
- H3K27ac
- H3K4me1
- H3K4me3
- CTCF
- TFBS
- TSS
- Promoter
- Enhancer
- SuperEnhancer
- WeakEnhancer
- Repressed
- UTR_5
- UTR_3
算是种类非常多了,如果对精度没有要求,就可以直接用了,全部是bed格式的。
参考:
Chromatin accessibility and the regulatory epigenome
Identifying and mitigating bias in next-generation sequencing methods for chromatin biology - 刘小乐
Chromatin Structure Research Methods
Introduction to ChIP-seq and ATAC-seq - 非常赞
Mapping DNA-protein interactions via ChIP-seq - 非常详细
如何通过CHIP-seq分析鉴别基因启动子和增强子 - ChIP-seq详解
ChIP-seq实践(H3K27Ac,enhancer的筛选和enhancer相关基因的GO分析) - 实战


【推荐】国内首个AI IDE,深度理解中文开发场景,立即下载体验Trae
【推荐】编程新体验,更懂你的AI,立即体验豆包MarsCode编程助手
【推荐】抖音旗下AI助手豆包,你的智能百科全书,全免费不限次数
【推荐】轻量又高性能的 SSH 工具 IShell:AI 加持,快人一步
· 从 HTTP 原因短语缺失研究 HTTP/2 和 HTTP/3 的设计差异
· AI与.NET技术实操系列:向量存储与相似性搜索在 .NET 中的实现
· 基于Microsoft.Extensions.AI核心库实现RAG应用
· Linux系列:如何用heaptrack跟踪.NET程序的非托管内存泄露
· 开发者必知的日志记录最佳实践
· winform 绘制太阳,地球,月球 运作规律
· AI与.NET技术实操系列(五):向量存储与相似性搜索在 .NET 中的实现
· 超详细:普通电脑也行Windows部署deepseek R1训练数据并当服务器共享给他人
· 【硬核科普】Trae如何「偷看」你的代码?零基础破解AI编程运行原理
· 上周热点回顾(3.3-3.9)