深度学习与医学图像处理 案例学习3——Covid19肺炎检测(CXR图像)
内容引用自https://www.kaggle.com/mahmoudreda55/x-ray-covid19-95
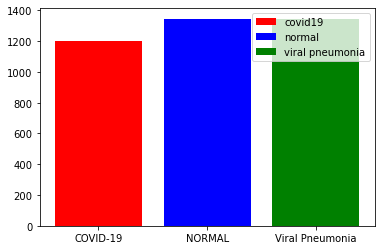
数据集中的影像模式为胸部X线,共有COVID-19数据1200例,1341例正常人影像和1345例病毒性肺炎影像。
#导入数据包
import kerastuner as kt from tensorflow import keras import tensorflow as tf from kerastuner.tuners import RandomSearch from kerastuner.engine.hyperparameters import HyperParameter as hp from keras.layers import Dense,Dropout,Activation,Add,MaxPooling2D,Conv2D,Flatten from keras.models import Sequential from keras.preprocessing.image import ImageDataGenerator import numpy as np import matplotlib.pyplot as plt from keras.applications import VGG19 from keras import layers from keras.preprocessing import image import matplotlib.pyplot as plt import seaborn as sns import os
import numpy as np
import pandas as pd
#读入数据
base_path='/kaggle/input/covid19-radiography-database/COVID-19 Radiography Database'
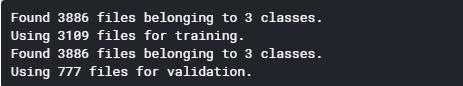
train_data = tf.keras.preprocessing.image_dataset_from_directory( base_path, labels="inferred", label_mode="int", class_names=None, color_mode="rgb", batch_size=32, image_size=(244, 244), shuffle=True, seed=123, validation_split=0.2, subset="training", interpolation="bilinear", follow_links=False, ) val_data = tf.keras.preprocessing.image_dataset_from_directory( base_path, labels="inferred", label_mode="int", class_names=None, color_mode="rgb", batch_size=32, image_size=(244, 244), shuffle=True, seed=123, validation_split=0.2, subset="validation", interpolation="bilinear", follow_links=False, )
#该方法适合整理数据,代码比较简单
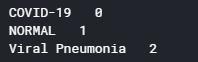
#分类 class_names = ["COVID-19","NORMAL","Viral Pneumonia"] for i in range(len(class_names)): print(class_names[i] ," " , i)
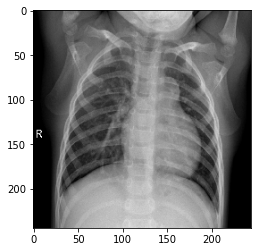
#显示一副病毒性肺炎的图片
image_path=os.path.join(base_path,'Viral Pneumonia/Viral Pneumonia (1024).png') new_img=image.load_img(image_path,target_size=(244,244)) print('type of new_img',type(new_img)) img = image.img_to_array(new_img) print('shape of img',img.shape) img = np.expand_dims(img, axis=0) print('shape of img',img.shape) print("Viral Pneumonia") plt.imshow(new_img)

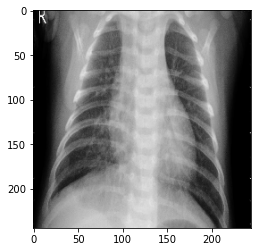
#显示一例正常人的数据
image_path=os.path.join(base_path,'NORMAL/NORMAL (1013).png') new_img = image.load_img(image_path, target_size=(244, 244)) img = image.img_to_array(new_img) img = np.expand_dims(img, axis=0) print("NORMAL") plt.imshow(new_img)
#显示1例新冠
image_path=os.path.join(base_path,'COVID/COVID (123).png') new_img = image.load_img(image_path, target_size=(244, 244)) img = image.img_to_array(new_img) img = np.expand_dims(img, axis=0) print("COVID-19") plt.imshow(new_img)
covid_num=len(os.listdir(base_path+'/COVID')) normal_num=len(os.listdir(base_path+'/NORMAL')) pneumonia_num=len(os.listdir(base_path+'/Viral Pneumonia'))
#查看三个数据集对比
fig, ax = plt.subplots() ax.bar("COVID-19",covid_num,color="r",label="covid19") ax.bar("NORMAL",normal_num,color="b",label="normal") ax.bar("Viral Pneumonia",pneumonia_num,color="g",label="viral pneumonia") ax.legend()
#CNN模型
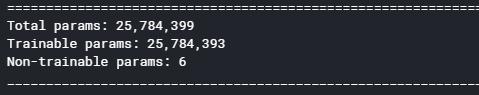
model = tf.keras.models.Sequential([ layers.BatchNormalization(), #244,244,3 layers.Conv2D(32, 3, activation='relu'), #242,242,32 layers.MaxPooling2D(), #121,121,32 layers.Conv2D(64, 3, activation='relu'), #119,119,64 layers.MaxPooling2D(), #59,59,64 layers.Dropout(0.3), #59,59,64 layers.Conv2D(128, 3, activation='relu'), #57,57,128 layers.MaxPooling2D(), #28,28,128 layers.Dropout(0.2), #28,28,128 layers.Flatten(), #100352 layers.Dense(256, activation='relu'), #256 layers.Dropout(0.15), layers.Dense(3, activation= 'softmax') #771 ])
model.summary()
#提前停止设置,5个epoch验证损失函数无提升则终止
early = tf.keras.callbacks.EarlyStopping(monitor='val_loss',patience=5)
model.compile(optimizer='adam',loss='sparse_categorical_crossentropy', metrics=['accuracy'])
history=model.fit(train_data,validation_data=val_data,batch_size=32,epochs = 10,callbacks=[early])

#显示结果
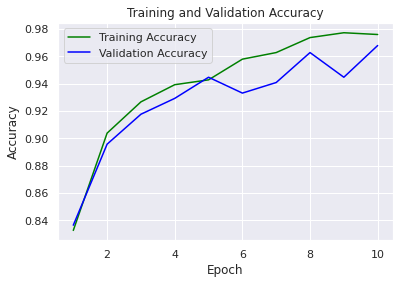
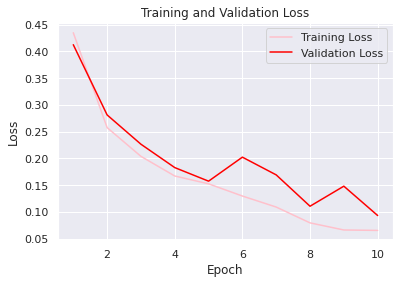
sns.set() acc = history.history['accuracy'] val_acc = history.history['val_accuracy'] loss = history.history['loss'] val_loss = history.history['val_loss'] epochs = range(1, len(loss) + 1) #accuracy plot plt.plot(epochs, acc, color='green', label='Training Accuracy') plt.plot(epochs, val_acc, color='blue', label='Validation Accuracy') plt.title('Training and Validation Accuracy') plt.ylabel('Accuracy') plt.xlabel('Epoch') plt.legend() plt.figure() #loss plot plt.plot(epochs, loss, color='pink', label='Training Loss') plt.plot(epochs, val_loss, color='red', label='Validation Loss') plt.title('Training and Validation Loss') plt.xlabel('Epoch') plt.ylabel('Loss') plt.legend() plt.show()
#使用前面的例子看结果 image_path=os.path.join(base_path,'COVID/COVID (123).png') new_img = image.load_img(image_path, target_size=(244, 244)) img = image.img_to_array(new_img) img = np.expand_dims(img, axis=0) prediction = model.predict(img) prediction = np.argmax(prediction,axis=1) print(prediction) print(class_names[prediction[0]])

image_path=os.path.join(base_path,'NORMAL/NORMAL (1013).png') new_img = image.load_img(image_path, target_size=(244, 244)) img = image.img_to_array(new_img) img = np.expand_dims(img, axis=0) prediction = model.predict(img) prediction = np.argmax(prediction,axis=1) print(prediction) print(class_names[prediction[0]])

image_path=os.path.join(base_path,'Viral Pneumonia/Viral Pneumonia (1024).png') new_img = image.load_img(image_path, target_size=(244, 244)) img = image.img_to_array(new_img) img = np.expand_dims(img, axis=0) prediction = model.predict(img) prediction = np.argmax(prediction,axis=1) print(prediction) print(class_names[prediction[0]])

#保存模型
model.save('model.h5')