plot a critical difference diagram , MATLAB code
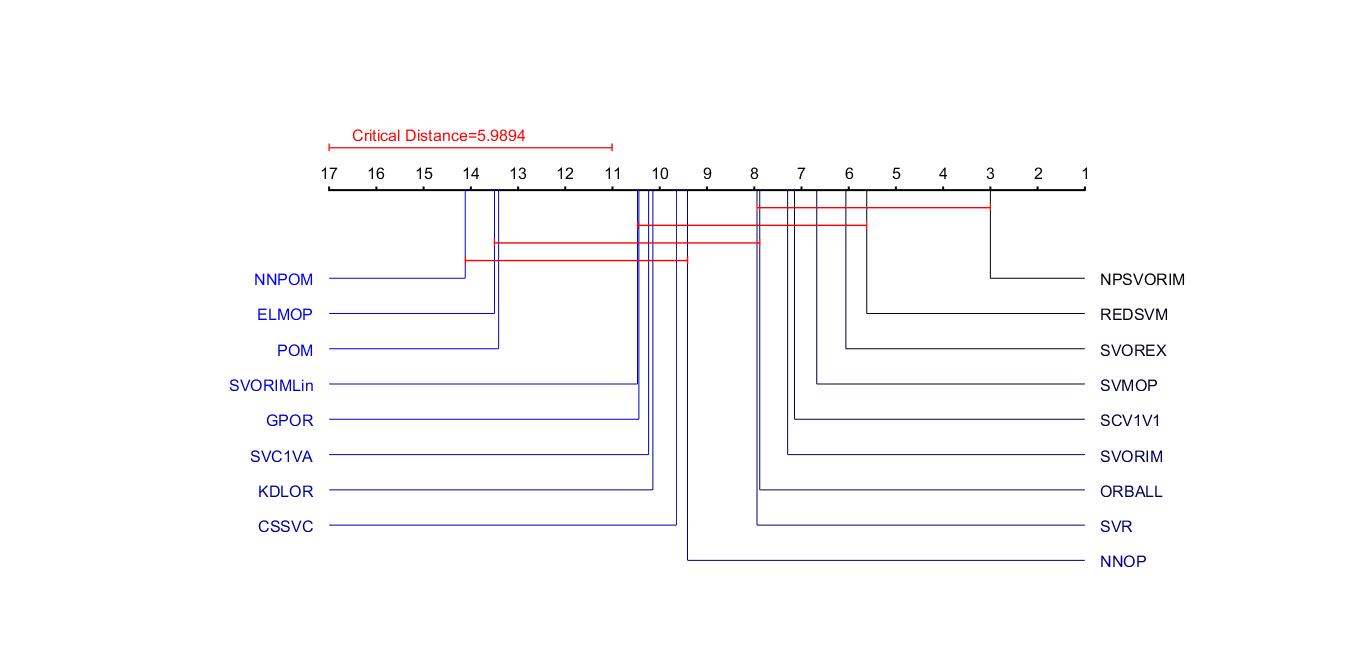
建立criticaldifference函数
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 | function cd = criticaldifference(s,labels,alpha)%% CRITICALDIFFERNCE - plot a critical difference diagram%% CRITICALDIFFERENCE(S,LABELS) produces a critical difference diagram [1]% displaying the statistical significance (or otherwise) of a matrix of% scores, S, achieved by a set of machine learning algorithms. Here% LABELS is a cell array of strings giving the name of each algorithm.%% References% % [1] Demsar, J., "Statistical comparisons of classifiers over multiple% datasets", Journal of Machine Learning Research, vol. 7, pp. 1-30,% 2006.%%% File : criticaldifference.m%% Date : Monday 14th April 2008%% Author : Gavin C. Cawley%% Description : Sparse multinomial logistic regression using a Laplace prior.%% History : 14/04/2008 - v1.00%% Copyright : (c) Dr Gavin C. Cawley, April 2008.%% This program is free software; you can redistribute it and/or modify% it under the terms of the GNU General Public License as published by% the Free Software Foundation; either version 2 of the License, or% (at your option) any later version.%% This program is distributed in the hope that it will be useful,% but WITHOUT ANY WARRANTY; without even the implied warranty of% MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the% GNU General Public License for more details.%% You should have received a copy of the GNU General Public License% along with this program; if not, write to the Free Software% Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA%% Thanks to Gideon Dror for supplying the extended table of critical values.if nargin < 3 alpha = 0.1;end% convert scores into ranks [N,k] = size(s);[S,r] = sort(s');idx = k*repmat(0:N-1, k, 1)' + r';R = repmat(1:k, N, 1);S = S';for i=1:N for j=1:k index = S(i,j) == S(i,:); R(i,index) = mean(R(i,index)); endendr(idx) = R;r = r';% compute critical differenceif alpha == 0.01 qalpha = [0.000 2.576 2.913 3.113 3.255 3.364 3.452 3.526 3.590 3.646 ... 3.696 3.741 3.781 3.818 3.853 3.884 3.914 3.941 3.967 3.992 ... 4.015 4.037 4.057 4.077 4.096 4.114 4.132 4.148 4.164 4.179 ... 4.194 4.208 4.222 4.236 4.249 4.261 4.273 4.285 4.296 4.307 ... 4.318 4.329 4.339 4.349 4.359 4.368 4.378 4.387 4.395 4.404 ... 4.412 4.420 4.428 4.435 4.442 4.449 4.456 ]; elseif alpha == 0.05 qalpha = [0.000 1.960 2.344 2.569 2.728 2.850 2.948 3.031 3.102 3.164 ... 3.219 3.268 3.313 3.354 3.391 3.426 3.458 3.489 3.517 3.544 ... 3.569 3.593 3.616 3.637 3.658 3.678 3.696 3.714 3.732 3.749 ... 3.765 3.780 3.795 3.810 3.824 3.837 3.850 3.863 3.876 3.888 ... 3.899 3.911 3.922 3.933 3.943 3.954 3.964 3.973 3.983 3.992 ... 4.001 4.009 4.017 4.025 4.032 4.040 4.046]; elseif alpha == 0.1 qalpha = [0.000 1.645 2.052 2.291 2.460 2.589 2.693 2.780 2.855 2.920 ... 2.978 3.030 3.077 3.120 3.159 3.196 3.230 3.261 3.291 3.319 ... 3.346 3.371 3.394 3.417 3.439 3.459 3.479 3.498 3.516 3.533 ... 3.550 3.567 3.582 3.597 3.612 3.626 3.640 3.653 3.666 3.679 ... 3.691 3.703 3.714 3.726 3.737 3.747 3.758 3.768 3.778 3.788 ... 3.797 3.806 3.814 3.823 3.831 3.838 3.846];else error('alpha must be 0.01, 0.05 or 0.1');endcd = qalpha(k)*sqrt(k*(k+1)/(6*N));figure(1);clfaxis offaxis([-0.2 1.2 -20 140]);axis xy tics = repmat((0:(k-1))/(k-1), 3, 1);line(tics(:), repmat([100, 101, 100], 1, k), 'LineWidth', 1.5, 'Color', 'k');%tics = repmat(((0:(k-2))/(k-1)) + 0.5/(k-1), 3, 1);%line(tics(:), repmat([100, 101, 100], 1, k-1), 'LineWidth', 1.5, 'Color', 'k');line([0 0 0 cd/(k-1) cd/(k-1) cd/(k-1)], [113 111 112 112 111 113], 'LineWidth', 1, 'Color', 'r');text(0.03, 116, ['Critical Distance=' num2str(cd)], 'FontSize', 12, 'HorizontalAlignment', 'left', 'Color', 'r');for i=1:k text((i-1)/(k-1), 105, num2str(k-i+1), 'FontSize', 12, 'HorizontalAlignment', 'center');end% compute average ranksr = mean(r);[r,idx] = sort(r);% compute statistically similar cliquesclique = repmat(r,k,1) - repmat(r',1,k);clique(clique<0) = realmax; clique = clique < cd;for i=k:-1:2 if all(clique(i-1,clique(i,:))==clique(i,clique(i,:))) clique(i,:) = 0; endendn = sum(clique,2);clique = clique(n>1,:);n = size(clique,1);%yanse={'b','g','y','m','r'};b=linspace(0,1,k);% labels displayed on the rightfor i=1:ceil(k/2) line([(k-r(i))/(k-1) (k-r(i))/(k-1) 1], [100 100-3*(n+1)-10*i 100-3*(n+1)-10*i], 'Color', [0 0 b(i)]); %text(1.2, 100 - 5*(n+1)- 10*i + 2, num2str(r(i)), 'FontSize', 10, 'HorizontalAlignment', 'right'); text(1.02, 100 - 3*(n+1) - 10*i, labels{idx(i)}, 'FontSize', 12, 'VerticalAlignment', 'middle', 'HorizontalAlignment', 'left', 'Color', [0 0 b(i)]);end% labels displayed on the leftfor i=ceil(k/2)+1:k line([(k-r(i))/(k-1) (k-r(i))/(k-1) 0], [100 100-3*(n+1)-10*(k-i+1) 100-3*(n+1)-10*(k-i+1)], 'Color', [0 0 b(i)]); %text(-0.2, 100 - 5*(n+1) -10*(k-i+1)+2, num2str(r(i)), 'FontSize', 10, 'HorizontalAlignment', 'left'); text(-0.02, 100 - 3*(n+1) -10*(k-i+1), labels{idx(i)}, 'FontSize', 12, 'VerticalAlignment', 'middle', 'HorizontalAlignment', 'right', 'Color', [0 0 b(i)]);end% group cliques of statistically similar classifiersfor i=1:size(clique,1) R = r(clique(i,:)); %line([((k-min(R))/(k-1)) + 0.015 ((k - max(R))/(k-1)) - 0.015], [100-5*i 100-5*i], 'LineWidth', 1, 'Color', 'r'); %line([0 0 0 cd/(k-1) cd/(k-1) cd/(k-1)], [113 111 112 112 111 113], 'LineWidth', 1, 'Color', 'r'); line([((k-min(R))/(k-1)) ((k-min(R))/(k-1)) ((k-min(R))/(k-1)) ((k - max(R))/(k-1)) ((k - max(R))/(k-1)) ((k - max(R))/(k-1))], [100+1-5*i 100-1-5*i 100-5*i 100-5*i 100-1-5*i 100+1-5*i], 'LineWidth', 1, 'Color', 'r');end |
可执行m文件:
1 2 3 4 5 6 7 | load Datas=AccMatrix;labels={'SCV1V1','SVC1VA','SVR','CSSVC','SVMOP','NNOP','ELMOP','POM',... 'NNPOM', 'SVOREX','SVORIM','SVORIMLin','KDLOR','GPOR','REDSVM','ORBALL' ,'NPSVORIM'};%方法的标签alpha=0.05; %显著性水平0.1,0.05或0.01cd = criticaldifference(s,labels,alpha) |
AccMatrix=[
0.28 0.12 0.28 0.11 0.32 0.08 0.26 0.13 0.37 0.10 0.28 0.12 0.42 0.21 0.38 0.17 0.36 0.14 0.36 0.13 0.38 0.12 0.37 0.10 0.34 0.15 0.39 0.09 0.37 0.12 0.36 0.13 0.37 0.11
0.31 0.12 0.33 0.11 0.34 0.13 0.32 0.11 0.32 0.09 0.24 0.11 0.40 0.18 0.50 0.15 0.34 0.18 0.35 0.12 0.34 0.12 0.34 0.12 0.33 0.11 0.48 0.17 0.33 0.11 0.30 0.12 0.28 0.14
0.36 0.09 0.40 0.14 0.39 0.11 0.39 0.13 0.40 0.09 0.39 0.11 0.44 0.16 0.62 0.15 0.50 0.13 0.37 0.13 0.37 0.13 0.37 0.13 0.39 0.12 0.55 0.10 0.38 0.13 0.36 0.12 0.32 0.10
0.22 0.12 0.28 0.16 0.24 0.10 0.27 0.15 0.27 0.11 0.29 0.11 0.39 0.13 0.65 0.14 0.39 0.14 0.26 0.11 0.27 0.11 0.32 0.11 0.26 0.11 0.36 0.16 0.27 0.12 0.30 0.10 0.22 0.10
0.44 0.06 0.45 0.06 0.40 0.07 0.43 0.07 0.46 0.06 0.41 0.06 0.44 0.08 0.50 0.08 0.45 0.09 0.41 0.07 0.40 0.07 0.48 0.07 0.43 0.05 0.67 0.04 0.40 0.07 0.40 0.06 0.41 0.05
0.03 0.03 0.04 0.03 0.04 0.02 0.04 0.02 0.04 0.03 0.04 0.02 0.06 0.02 0.03 0.02 0.03 0.03 0.03 0.02 0.03 0.02 0.03 0.02 0.03 0.02 0.03 0.02 0.03 0.02 0.04 0.03 0.03 0.03
0.03 0.01 0.03 0.01 0.16 0.03 0.03 0.01 0.03 0.01 0.04 0.01 0.09 0.02 0.09 0.02 0.06 0.05 0.00 0.01 0.00 0.01 0.09 0.02 0.16 0.03 0.03 0.01 0.00 0.00 0.03 0.02 0.02 0.01
0.42 0.03 0.44 0.03 0.43 0.03 0.43 0.03 0.42 0.03 0.42 0.03 0.43 0.02 0.43 0.03 0.46 0.03 0.43 0.03 0.43 0.03 0.43 0.03 0.51 0.03 0.42 0.03 0.43 0.03 0.44 0.03 0.43 0.03
0.01 0.00 0.01 0.01 0.03 0.01 0.01 0.01 0.00 0.00 0.03 0.01 0.16 0.01 0.84 0.30 0.11 0.02 0.01 0.01 0.01 0.01 0.08 0.01 0.05 0.01 0.04 0.01 0.01 0.00 0.01 0.01 0.01 0.00
0.43 0.04 0.43 0.06 0.46 0.07 0.43 0.06 0.45 0.10 0.53 0.09 0.57 0.13 0.66 0.16 0.62 0.14 0.45 0.06 0.45 0.07 0.43 0.08 0.47 0.09 0.42 0.03 0.44 0.05 0.46 0.09 0.42 0.08
0.05 0.03 0.05 0.03 0.07 0.04 0.05 0.03 0.07 0.03 0.06 0.03 0.07 0.03 0.71 0.03 0.06 0.03 0.02 0.01 0.02 0.01 0.74 0.01 0.11 0.03 0.05 0.02 0.02 0.01 0.05 0.02 0.04 0.03
0.36 0.03 0.45 0.03 0.36 0.03 0.44 0.03 0.35 0.03 0.42 0.04 0.43 0.03 0.85 0.02 0.46 0.04 0.36 0.03 0.36 0.03 0.36 0.02 0.37 0.03 0.31 0.03 0.36 0.03 0.38 0.03 0.34 0.03
0.37 0.02 0.37 0.02 0.38 0.02 0.37 0.02 0.37 0.02 0.37 0.03 0.37 0.02 0.38 0.03 0.38 0.02 0.38 0.02 0.38 0.02 0.39 0.02 0.46 0.03 0.39 0.03 0.37 0.02 0.39 0.03 0.37 0.03
0.25 0.06 0.26 0.06 0.32 0.07 0.27 0.06 0.26 0.04 0.39 0.06 0.38 0.06 0.53 0.19 0.55 0.08 0.32 0.05 0.32 0.07 0.41 0.07 0.30 0.07 0.39 0.07 0.32 0.07 0.29 0.05 0.27 0.05
0.35 0.02 0.36 0.02 0.37 0.02 0.36 0.02 0.36 0.02 0.40 0.02 0.40 0.02 0.40 0.02 0.40 0.02 0.37 0.02 0.37 0.02 0.41 0.02 0.35 0.02 0.39 0.01 0.37 0.02 0.33 0.02 0.36 0.02
0.31 0.04 0.33 0.03 0.30 0.03 0.32 0.03 0.29 0.03 0.31 0.04 0.30 0.04 0.29 0.03 0.34 0.13 0.29 0.03 0.28 0.03 0.29 0.04 0.36 0.03 0.29 0.03 0.29 0.03 0.32 0.02 0.29 0.03
0.74 0.02 0.82 0.03 0.75 0.02 0.80 0.03 0.74 0.02 0.71 0.02 0.75 0.02 0.74 0.02 0.73 0.03 0.71 0.03 0.75 0.02 0.76 0.02 0.81 0.03 0.71 0.03 0.75 0.02 0.76 0.02 0.75 0.03 ];





【推荐】国内首个AI IDE,深度理解中文开发场景,立即下载体验Trae
【推荐】编程新体验,更懂你的AI,立即体验豆包MarsCode编程助手
【推荐】抖音旗下AI助手豆包,你的智能百科全书,全免费不限次数
【推荐】轻量又高性能的 SSH 工具 IShell:AI 加持,快人一步
· 开发者必知的日志记录最佳实践
· SQL Server 2025 AI相关能力初探
· Linux系列:如何用 C#调用 C方法造成内存泄露
· AI与.NET技术实操系列(二):开始使用ML.NET
· 记一次.NET内存居高不下排查解决与启示
· 阿里最新开源QwQ-32B,效果媲美deepseek-r1满血版,部署成本又又又降低了!
· 开源Multi-agent AI智能体框架aevatar.ai,欢迎大家贡献代码
· Manus重磅发布:全球首款通用AI代理技术深度解析与实战指南
· 被坑几百块钱后,我竟然真的恢复了删除的微信聊天记录!
· 没有Manus邀请码?试试免邀请码的MGX或者开源的OpenManus吧