R_MarinStats_Notes3
5.1 simple linear regression in r
> plot(Age, LungCap, main="Scatterplot")
> cor(Age, LungCap)
[1] 0.8196749
>
> ?lm
> mod = lm(LungCap ~ Age)
> summary(mod)
Call:
lm(formula = LungCap ~ Age)
Residuals:
Min 1Q Median 3Q Max
-4.7799 -1.0203 -0.0005 0.9789 4.2650
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.14686 0.18353 6.249 7.06e-10 ***
Age 0.54485 0.01416 38.476 < 2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.526 on 723 degrees of freedom
Multiple R-squared: 0.6719, Adjusted R-squared: 0.6714
F-statistic: 1480 on 1 and 723 DF, p-value: < 2.2e-16
> attributes(mod)
$names
[1] "coefficients" "residuals" "effects" "rank" "fitted.values" "assign"
[7] "qr" "df.residual" "xlevels" "call" "terms" "model"
$class
[1] "lm"
> mod$coefficients
(Intercept) Age
1.1468578 0.5448484
> coef(mod)
(Intercept) Age
1.1468578 0.5448484
> plot(Age, LungCap, main="Scatterplot")
> abline(mod, col=2, lwd=3)
>
> confint(mod)
2.5 % 97.5 %
(Intercept) 0.7865454 1.5071702
Age 0.5170471 0.5726497
> confint(mod, level=0.99)
0.5 % 99.5 %
(Intercept) 0.6728686 1.6208470
Age 0.5082759 0.5814209
> summary(mod)
Call:
lm(formula = LungCap ~ Age)
Residuals:
Min 1Q Median 3Q Max
-4.7799 -1.0203 -0.0005 0.9789 4.2650
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.14686 0.18353 6.249 7.06e-10 ***
Age 0.54485 0.01416 38.476 < 2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.526 on 723 degrees of freedom
Multiple R-squared: 0.6719, Adjusted R-squared: 0.6714
F-statistic: 1480 on 1 and 723 DF, p-value: < 2.2e-16
> anova(mod)
Analysis of Variance Table
Response: LungCap
Df Sum Sq Mean Sq F value Pr(>F)
Age 1 3447.0 3447.0 1480.4 < 2.2e-16 ***
Residuals 723 1683.5 2.3
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
>
> sqrt(2.3)
[1] 1.516575
> # 1.516575 is equal to 1.526
MORE INFO:3
https://stackoverflow.com/questions/5587676/pull-out-p-values-and-r-squared-from-a-linear-regression
Notice that summary(fit) generates an object with all the information you need. The beta, se, t and p vectors are stored in it. Get the p-values by selecting the 4th column of the coefficients matrix (stored in the summary object):
summary(fit)$coefficients[,4]
summary(fit)$r.squaredTry str(summary(fit)) to see all the info that this object contains.
> mod <- lm(LungCap ~ Age)
> mod
Call:
lm(formula = LungCap ~ Age)
Coefficients:
(Intercept) Age
1.1469 0.5448
> summary(mod)
Call:
lm(formula = LungCap ~ Age)
Residuals:
Min 1Q Median 3Q Max
-4.7799 -1.0203 -0.0005 0.9789 4.2650
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.14686 0.18353 6.249 7.06e-10 ***
Age 0.54485 0.01416 38.476 < 2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.526 on 723 degrees of freedom
Multiple R-squared: 0.6719, Adjusted R-squared: 0.6714
F-statistic: 1480 on 1 and 723 DF, p-value: < 2.2e-16
> attributes(summary(mod))
$names
[1] "call" "terms" "residuals" "coefficients" "aliased" "sigma"
[7] "df" "r.squared" "adj.r.squared" "fstatistic" "cov.unscaled"
$class
[1] "summary.lm"
> summary(mod)$r.squared
[1] 0.6718669
> summary(mod)$adj.r.squared
[1] 0.6714131
> summary(mod)$coefficients[, 4]
(Intercept) Age
7.056380e-10 4.077172e-177
> summary(mod)$coefficients[,1:4]
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.1468578 0.18352850 6.248936 7.056380e-10
Age 0.5448484 0.01416087 38.475634 4.077172e-177
> class(summary(mod))
[1] "summary.lm"
> class(summary(mod)$coefficients)
[1] "matrix"
> class(summary(mod)$coefficients[,4])
[1] "numeric"
> class(summary(mod)$coefficients[,1:4])
[1] "matrix"
>
> summary(mod)$coefficients[, 4]["(Intercept)"]
(Intercept)
7.05638e-10
> summary(mod)$coefficients[, 4]["Age"]
Age
4.077172e-177
>
> summary(mod)$coefficients[, 4][1]
(Intercept)
7.05638e-10
> summary(mod)$coefficients[, 4][2]
Age
4.077172e-177
> summary(mod)$coefficients[, 4][3]
<NA>
NA
Interpreting output from anova() when using lm() as input:
https://stats.stackexchange.com/questions/115304/interpreting-output-from-anova-when-using-lm-as-input
https://stats.stackexchange.com/questions/5135/interpretation-of-rs-lm-output
The anova() function call returns an ANOVA table. You can use it to get an ANOVA table any time you want one. Thus, the question becomes, 'why might I want an ANOVA table when I can just get tt-tests of my variables with standard output (i.e., the summary.lm() command)?'
First of all, you may be perfectly satisfied with the summary output, and that's fine. However, the ANOVA table may offer some advantages. First, if you have a categorical / factor variable with more than two levels, the summary output is hard to interpret. It will give you tests of individual levels against the reference level, but won't give you a test of the factor as a whole. Consider:
set.seed(8867) # this makes the example exactly reproducible
y = c(rnorm(10, mean=0, sd=1),
rnorm(10, mean=-.5, sd=1),
rnorm(10, mean=.5, sd=1) )
g = rep(c("A", "B", "C"), each=10)
model = lm(y~g)
summary(model)
# ...
# Residuals:
# Min 1Q Median 3Q Max
# -2.59080 -0.54685 0.04124 0.79890 2.56064
#
# Coefficients:
# Estimate Std. Error t value Pr(>|t|)
# (Intercept) -0.4440 0.3855 -1.152 0.260
# gB -0.9016 0.5452 -1.654 0.110
# gC 0.6729 0.5452 1.234 0.228
#
# Residual standard error: 1.219 on 27 degrees of freedom
# Multiple R-squared: 0.2372, Adjusted R-squared: 0.1807
# F-statistic: 4.199 on 2 and 27 DF, p-value: 0.02583
anova(model)
# Analysis of Variance Table
#
# Response: y
# Df Sum Sq Mean Sq F value Pr(>F)
# g 2 12.484 6.2418 4.199 0.02583 *
# Residuals 27 40.135 1.4865
Another reason you might prefer to look at an ANOVA table is that it allows you to use information about the possible associations between your independent variables and your dependent variable that gets thrown away by the tt-tests in the summary output. Consider your own example, you may notice that the pp-values from the two don't match (e.g., for v1, the pp-value in the summary output is 0.93732, but in the ANOVA table it's 0.982400). The reason is that your variables are not perfectly uncorrelated:
cor(my_data) # v1 v2 v3 response # v1 1.00000000 -0.23760679 -0.1312995 -0.00357923 # v2 -0.23760679 1.00000000 -0.2358402 0.06069167 # v3 -0.13129952 -0.23584024 1.0000000 0.32818751 # response -0.00357923 0.06069167 0.3281875 1.00000000
The result of this is that there are sums of squares that could be attributed to more than one of the variables. The tt-tests are equivalent to 'type III' tests of the sums of squares, but other tests are possible. The default ANOVA table uses 'type I' sums of squares, which can allow you to make more precise--and more powerful--tests of your hypotheses.
总离差平方和:SST(Sum of Squares for total) = TSS(total sum of squares)
SSE+SSR=SST RSS+ESS=TSS
> plot(mod) Hit <Return> to see next plot: Hit <Return> to see next plot: Hit <Return> to see next plot: Hit <Return> to see next plot: > par(mfrow=c(2,2)) > plot(mod) > par(mfrow=c(1,1)) > plot(mod) Hit <Return> to see next plot: Hit <Return> to see next plot: Hit <Return> to see next plot: Hit <Return> to see next plot:
5.3 multiple linear regression
> model1 <- lm(LungCap ~ Age+Height)
> summary(model1)
Call:
lm(formula = LungCap ~ Age + Height)
Residuals:
Min 1Q Median 3Q Max
-3.4080 -0.7097 -0.0078 0.7167 3.1679
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -11.747065 0.476899 -24.632 < 2e-16 ***
Age 0.126368 0.017851 7.079 3.45e-12 ***
Height 0.278432 0.009926 28.051 < 2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.056 on 722 degrees of freedom
Multiple R-squared: 0.843, Adjusted R-squared: 0.8425
F-statistic: 1938 on 2 and 722 DF, p-value: < 2.2e-16
> anova(model1)
Analysis of Variance Table
Response: LungCap
Df Sum Sq Mean Sq F value Pr(>F)
Age 1 3447.0 3447.0 3089.42 < 2.2e-16 ***
Height 1 877.9 877.9 786.84 < 2.2e-16 ***
Residuals 722 805.6 1.1
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
> cor(Age, Height, method="pearson")
[1] 0.8357368
> confint(model1, level=0.95)
2.5 % 97.5 %
(Intercept) -12.68333877 -10.8107918
Age 0.09132215 0.1614142
Height 0.25894454 0.2979192
> model2<- lm(LungCap ~ Age+Height+Smoke+Gender+Caesarean)
> summary(model2)
Call:
lm(formula = LungCap ~ Age + Height + Smoke + Gender + Caesarean)
Residuals:
Min 1Q Median 3Q Max
-3.3388 -0.7200 0.0444 0.7093 3.0172
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -11.32249 0.47097 -24.041 < 2e-16 ***
Age 0.16053 0.01801 8.915 < 2e-16 ***
Height 0.26411 0.01006 26.248 < 2e-16 ***
Smokeyes -0.60956 0.12598 -4.839 1.60e-06 ***
Gendermale 0.38701 0.07966 4.858 1.45e-06 ***
Caesareanyes -0.21422 0.09074 -2.361 0.0185 *
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.02 on 719 degrees of freedom
Multiple R-squared: 0.8542, Adjusted R-squared: 0.8532
F-statistic: 842.8 on 5 and 719 DF, p-value: < 2.2e-16
> plot(model2)
Hit <Return> to see next plot:
Hit <Return> to see next plot:
Hit <Return> to see next plot:
Hit <Return> to see next plot:
5.4 converting a numeric variable to a categorical variable in r
useful for making some cross-tabulations for a variable, or fitting a regression model when the linearity assumption is not valid for the variable
> ?cut
> #A <=50, B(50,55], C(55,60], D(60,65], E(65,70], F>70
> CatHeight <- cut(Height, breaks = c(0,50,55,60,65,70,100), labels=c("A","B","C","D","E","F"))
> Height[1:10]
[1] 62.1 74.7 69.7 71.0 56.9 58.7 63.3 70.4 70.5 59.2
> CatHeight[1:10]
[1] D F E F C C D F F C
Levels: A B C D E F
> #right = False [a, b)
> CatHeight <- cut(Height, breaks = c(0,50,55,60,65,70,100), labels=c("A","B","C","D","E","F"), right = F)
> Height[1:10]
[1] 62.1 74.7 69.7 71.0 56.9 58.7 63.3 70.4 70.5 59.2
> CatHeight[1:10]
[1] D F E F C C D F F C
Levels: A B C D E F
> CatHeight <- cut(Height, breaks = c(0,50,55,60,65,70,100), right = F)
> CatHeight[1:10]
[1] [60,65) [70,100) [65,70) [70,100) [55,60) [55,60) [60,65) [70,100) [70,100) [55,60)
Levels: [0,50) [50,55) [55,60) [60,65) [65,70) [70,100)
> CatHeight <- cut(Height, breaks = 4, right = F)
> CatHeight[1:10]
[1] [54.4,63.5) [72.7,81.8) [63.5,72.7) [63.5,72.7) [54.4,63.5) [54.4,63.5) [54.4,63.5) [63.5,72.7)
[9] [63.5,72.7) [54.4,63.5)
Levels: [45.3,54.4) [54.4,63.5) [63.5,72.7) [72.7,81.8)
5.5 dummy variable or indicator variables in r
> CatHeight <- cut(Height, breaks = c(0,50,55,60,65,70,100), labels=c("A","B","C","D","E","F"), right = F)
> CatHeight[1:10]
[1] D F E F C C D F F C
Levels: A B C D E F
> levels(Smoke)
[1] "no" "yes"
> #2 levels require 1 indicator variable to represent smoking status
>
> levels(CatHeight)
[1] "A" "B" "C" "D" "E" "F"
> mean(LungCap[CatHeight == "A"])
[1] 2.148611
> mean(LungCap[CatHeight == "B"])
[1] 3.65464
> mean(LungCap[CatHeight == "C"])
[1] 5.396983
> mean(LungCap[CatHeight == "D"])
[1] 7.170552
> mean(LungCap[CatHeight == "E"])
[1] 8.692063
> mean(LungCap[CatHeight == "F"])
[1] 10.80278
> mod <- lm(LungCap ~ CatHeight)
> summary(mod)
Call:
lm(formula = LungCap ~ CatHeight)
Residuals:
Min 1Q Median 3Q Max
-3.9671 -0.7720 -0.0456 0.7972 3.8722
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.1486 0.2929 7.336 5.97e-13 ***
CatHeightB 1.5060 0.3416 4.409 1.20e-05 ***
CatHeightC 3.2484 0.3148 10.319 < 2e-16 ***
CatHeightD 5.0219 0.3086 16.271 < 2e-16 ***
CatHeightE 6.5435 0.3065 21.347 < 2e-16 ***
CatHeightF 8.6542 0.3065 28.233 < 2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.243 on 719 degrees of freedom
Multiple R-squared: 0.7836, Adjusted R-squared: 0.7821
F-statistic: 520.7 on 5 and 719 DF, p-value: < 2.2e-16
5.6 Changing baseline(Reference) Category for a Categorical Variable in a linear regression model in r
By default, the category that R chooses to be the reference or baseline, is the first category that appears alphabetically or numerically. ('no' in this case)
> ?relevel
> mod1 <- lm(LungCap ~ Age + Smoke)
> summary(mod1)
Call:
lm(formula = LungCap ~ Age + Smoke)
Residuals:
Min 1Q Median 3Q Max
-4.8559 -1.0289 -0.0363 1.0083 4.1995
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.08572 0.18299 5.933 4.61e-09 ***
Age 0.55540 0.01438 38.628 < 2e-16 ***
Smokeyes -0.64859 0.18676 -3.473 0.000546 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.514 on 722 degrees of freedom
Multiple R-squared: 0.6773, Adjusted R-squared: 0.6764
F-statistic: 757.5 on 2 and 722 DF, p-value: < 2.2e-16
> table(Smoke)
Smoke
no yes
648 77
>
> Smoke <- relevel(Smoke, ref="yes")
> table(Smoke)
Smoke
yes no
77 648
> mod2 <- lm(LungCap ~ Age + Smoke)
> summary(mod2)
Call:
lm(formula = LungCap ~ Age + Smoke)
Residuals:
Min 1Q Median 3Q Max
-4.8559 -1.0289 -0.0363 1.0083 4.1995
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.43714 0.27375 1.597 0.110740
Age 0.55540 0.01438 38.628 < 2e-16 ***
Smokeno 0.64859 0.18676 3.473 0.000546 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.514 on 722 degrees of freedom
Multiple R-squared: 0.6773, Adjusted R-squared: 0.6764
F-statistic: 757.5 on 2 and 722 DF, p-value: < 2.2e-16
5.7 categorical variable into regression model in r
assumes there is no interaction between smoking and age
> model1 <- lm(LungCap ~ Age + Smoke)
> summary(model1)
Call:
lm(formula = LungCap ~ Age + Smoke)
Residuals:
Min 1Q Median 3Q Max
-4.8559 -1.0289 -0.0363 1.0083 4.1995
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.08572 0.18299 5.933 4.61e-09 ***
Age 0.55540 0.01438 38.628 < 2e-16 ***
Smokeyes -0.64859 0.18676 -3.473 0.000546 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.514 on 722 degrees of freedom
Multiple R-squared: 0.6773, Adjusted R-squared: 0.6764
F-statistic: 757.5 on 2 and 722 DF, p-value: < 2.2e-16
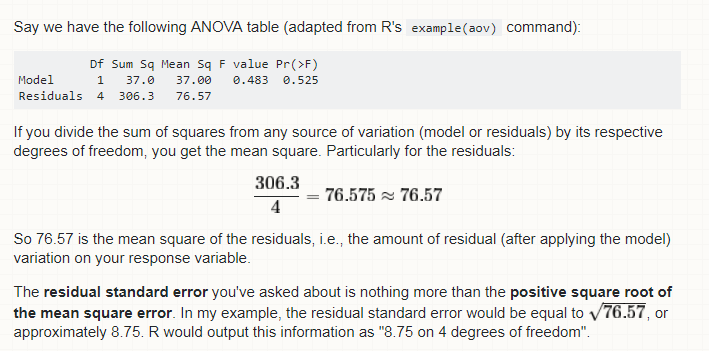
> plot(Age[Smoke=="no"], LungCap[Smoke=="no"], col="blue", ylim=c(0,15), xlab="Age", ylab="LungCap", main="LungCap vs. Age & Smoke")
> points(Age[Smoke=="yes"], LungCap[Smoke=="yes"], col="red", pch=16)
> legend(3, 15, legend = c("NonSmoker", "Smoker"), col=c("blue", "red"), pch=c(1,16), bty="n")
>
> abline(a=1.08, b=0.555, col="blue", lwd=3)
> abline(a=1.08-0.64859, b=0.555, col="red", lwd=3)
5.8 another example of including a categorical variable into a regression model in r
> levels(CatHeight)
[1] "A" "B" "C" "D" "E" "F"
> model2 <- lm(LungCap ~ Age + CatHeight)
> summary(model2)
Call:
lm(formula = LungCap ~ Age + CatHeight)
Residuals:
Min 1Q Median 3Q Max
-3.8492 -0.7711 0.0037 0.7449 3.4230
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.99913 0.29371 3.402 0.000707 ***
Age 0.19705 0.01868 10.546 < 2e-16 ***
CatHeightB 1.48107 0.31808 4.656 3.83e-06 ***
CatHeightC 2.67193 0.29819 8.961 < 2e-16 ***
CatHeightD 3.92646 0.30560 12.848 < 2e-16 ***
CatHeightE 5.01342 0.32019 15.658 < 2e-16 ***
CatHeightF 6.58093 0.34658 18.988 < 2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.157 on 718 degrees of freedom
Multiple R-squared: 0.8126, Adjusted R-squared: 0.811
F-statistic: 518.9 on 6 and 718 DF, p-value: < 2.2e-16
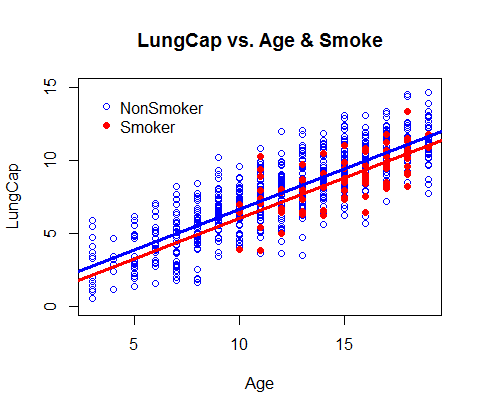
> plot(Age[CatHeight=="A"], LungCap[CatHeight=="A"], col=2, ylim = c(0,15), xlim=c(0,20), xlab="Age", ylab = "LungCap", main = "LungCap vs Age & CatHeight")
> points(Age[CatHeight=="B"], LungCap[CatHeight=="B"], col=3)
> points(Age[CatHeight=="C"], LungCap[CatHeight=="C"], col=4)
> points(Age[CatHeight=="D"], LungCap[CatHeight=="D"], col=5)
> points(Age[CatHeight=="E"], LungCap[CatHeight=="E"], col=6)
> points(Age[CatHeight=="F"], LungCap[CatHeight=="F"], col=7)
>
> legend(0, 15.5, legend=c("A","B","C","D","E","F"), col=c(2:7), pch=1, cex=0.8)
>
> abline(a=0.98, b=0.20, col =2, lwd=3)
> abline(a=2.46, b=0.20, col =3, lwd=3)
> abline(a=3.67, b=0.20, col =4, lwd=3)
> abline(a=4.92, b=0.20, col =5, lwd=3)
> abline(a=5.99, b=0.20, col =6, lwd=3)
> abline(a=7.52, b=0.20, col =7, lwd=3)
5.9 multiple linear regression with interaction in R
If x1 and x2 interact, this means that the effect of x1 on y depends on the value of x2, and vice versa
(the effect of age is modified by smoking)
> model1 <- lm(LungCap ~ Age * Smoke)
> coef(model1)
(Intercept) Age Smokeyes Age:Smokeyes
1.05157244 0.55823350 0.22601390 -0.05970463
> model1 <- lm(LungCap ~ Age + Smoke + Age:Smoke)
> summary(model1)
Call:
lm(formula = LungCap ~ Age + Smoke + Age:Smoke)
Residuals:
Min 1Q Median 3Q Max
-4.8586 -1.0174 -0.0251 1.0004 4.1996
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.05157 0.18706 5.622 2.7e-08 ***
Age 0.55823 0.01473 37.885 < 2e-16 ***
Smokeyes 0.22601 1.00755 0.224 0.823
Age:Smokeyes -0.05970 0.06759 -0.883 0.377
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.515 on 721 degrees of freedom
Multiple R-squared: 0.6776, Adjusted R-squared: 0.6763
F-statistic: 505.1 on 3 and 721 DF, p-value: < 2.2e-16
Age:Smokeyes is not significant, the interaction should not be included in the model
5.10 interaction between factors in a linear regression model
5.11 comparing models using the partial F-test in R
partial F-test is used in model building and variable selection to help decide if a variable or term can be removed from a model without making the model significantly worse (does it result in a statistically significant increase in the sum of the squared errors a.k.a. residual sum of squares?)
Full model, reduced modeld
partial F-test is used to test nested model
H0: no significant difference in sse of full and reduced models
H1: full model has a significantly lower sse than the reduced model
> Full.Model <- lm(LungCap ~ Age + I(Age^2))
> Reduced.Model <- lm(LungCap ~ Age)
> summary(Full.Model)
Call:
lm(formula = LungCap ~ Age + I(Age^2))
Residuals:
Min 1Q Median 3Q Max
-4.8446 -1.0150 -0.0212 1.0442 4.1942
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.610275 0.433197 1.409 0.159
Age 0.647707 0.076549 8.461 <2e-16 ***
I(Age^2) -0.004354 0.003184 -1.367 0.172
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.525 on 722 degrees of freedom
Multiple R-squared: 0.6727, Adjusted R-squared: 0.6718
F-statistic: 742 on 2 and 722 DF, p-value: < 2.2e-16
> summary(Reduced.Model)
Call:
lm(formula = LungCap ~ Age)
Residuals:
Min 1Q Median 3Q Max
-4.7799 -1.0203 -0.0005 0.9789 4.2650
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.14686 0.18353 6.249 7.06e-10 ***
Age 0.54485 0.01416 38.476 < 2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.526 on 723 degrees of freedom
Multiple R-squared: 0.6719, Adjusted R-squared: 0.6714
F-statistic: 1480 on 1 and 723 DF, p-value: < 2.2e-16
> anova(Reduced.Model, Full.Model)
Analysis of Variance Table
Model 1: LungCap ~ Age
Model 2: LungCap ~ Age + I(Age^2)
Res.Df RSS Df Sum of Sq F Pr(>F)
1 723 1683.5
2 722 1679.1 1 4.3476 1.8694 0.172
> #not necessary to include age^2
>
> model1 <- lm(LungCap ~ Age+Gender+Smoke+Height)
> model2 <- lm(LungCap ~ Age+Gender+Smoke)
> anova(model2, model1)
Analysis of Variance Table
Model 1: LungCap ~ Age + Gender + Smoke
Model 2: LungCap ~ Age + Gender + Smoke + Height
Res.Df RSS Df Sum of Sq F Pr(>F)
1 721 1467.79
2 720 753.57 1 714.22 682.39 < 2.2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
> #necessary to include Smoke
5.12 polynomial regression in r ( fitting and accessing polynomial models in r)
use residual plots to assess linearity and check other model assumptions
poly(Height, degree=2, raw = T)
degree =2 include Height and Height squared; if raw == F, R will use orthogonal polynomials
If we include Height^3, we must also include Height, Height^2
> LungCapData2 <- read.table(file.choose(), header = T, sep = ",")
> View(LungCapData2)
> detach(LungCapData)
> attach(LungCapData2)
The following object is masked _by_ .GlobalEnv:
Smoke
> Smoke[1:10]
[1] no yes no no no no no no no no
Levels: no yes
> rm(Smoke)
> Smoke[1:10]
[1] no no no no no no no no no no
Levels: no yes
> plot(Height, LungCap, main="Polynomial Regression", las=1)
> model1 <- lm(LungCap ~ Height)
> summary(model1)
Call:
lm(formula = LungCap ~ Height)
Residuals:
Min 1Q Median 3Q Max
-5.2550 -0.7986 -0.0120 0.7342 6.3581
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -18.298036 0.544380 -33.61 <2e-16 ***
Height 0.395927 0.008865 44.66 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.292 on 652 degrees of freedom
Multiple R-squared: 0.7537, Adjusted R-squared: 0.7533
F-statistic: 1995 on 1 and 652 DF, p-value: < 2.2e-16
> abline(model1, lwd=3, col="red")
> #use residual plots to check linearity assumption
> plot(model1)
Hit <Return> to see next plot:
Hit <Return> to see next plot:
Hit <Return> to see next plot:
Hit <Return> to see next plot:
> model2 <- lm(LungCap ~ Height + I(Height^2))
> summary(model2)
Call:
lm(formula = LungCap ~ Height + I(Height^2))
Residuals:
Min 1Q Median 3Q Max
-5.4031 -0.6878 -0.0076 0.6577 5.9910
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 16.080634 4.509553 3.566 0.000389 ***
Height -0.750147 0.149566 -5.015 6.83e-07 ***
I(Height^2) 0.009466 0.001233 7.675 6.07e-14 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.238 on 651 degrees of freedom
Multiple R-squared: 0.7741, Adjusted R-squared: 0.7734
F-statistic: 1115 on 2 and 651 DF, p-value: < 2.2e-16
>
> model2again <- lm(LungCap ~ poly(Height, degree=2, raw=T))
> summary(model2again)
Call:
lm(formula = LungCap ~ poly(Height, degree = 2, raw = T))
Residuals:
Min 1Q Median 3Q Max
-5.4031 -0.6878 -0.0076 0.6577 5.9910
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 16.080634 4.509553 3.566 0.000389 ***
poly(Height, degree = 2, raw = T)1 -0.750147 0.149566 -5.015 6.83e-07 ***
poly(Height, degree = 2, raw = T)2 0.009466 0.001233 7.675 6.07e-14 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 1.238 on 651 degrees of freedom
Multiple R-squared: 0.7741, Adjusted R-squared: 0.7734
F-statistic: 1115 on 2 and 651 DF, p-value: < 2.2e-16
> lines(smooth.spline(Height, predict(model2), col="blue", lwd=3))
Error in smooth.spline(Height, predict(model2), col = "blue", lwd = 3) :
unused arguments (col = "blue", lwd = 3)
> lines(smooth.spline(Height, predict(model2)), col="blue", lwd=3)
>
> anova(model1, model2)
Analysis of Variance Table
Model 1: LungCap ~ Height
Model 2: LungCap ~ Height + I(Height^2)
Res.Df RSS Df Sum of Sq F Pr(>F)
1 652 1088.41
2 651 998.09 1 90.314 58.907 6.069e-14 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
>
> model3 <- lm(LungCap ~ Height + I(Height^2) + I(Height^3))
> summary(model3)
Call:
lm(formula = LungCap ~ Height + I(Height^2) + I(Height^3))
Residuals:
Min 1Q Median 3Q Max
-5.3885 -0.6900 0.0069 0.6511 5.9936
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -6.293e-01 3.803e+01 -0.017 0.987
Height 9.179e-02 1.908e+00 0.048 0.962
I(Height^2) -4.567e-03 3.173e-02 -0.144 0.886
I(Height^3) 7.739e-05 1.749e-04 0.443 0.658
Residual standard error: 1.239 on 650 degrees of freedom
Multiple R-squared: 0.7742, Adjusted R-squared: 0.7731
F-statistic: 742.7 on 3 and 650 DF, p-value: < 2.2e-16
> lines(smooth.spline(Height, predict(model3)), col="green", lwd=3, lty=3)
> anova(model2, model3)
Analysis of Variance Table
Model 1: LungCap ~ Height + I(Height^2)
Model 2: LungCap ~ Height + I(Height^2) + I(Height^3)
Res.Df RSS Df Sum of Sq F Pr(>F)
1 651 998.09
2 650 997.79 1 0.30066 0.1959 0.6582

