sklearn.utils.class_weight.compute_class_weight
https://blog.csdn.net/FY_2018/article/details/116951278
compute_class_weight这个函数的作用是对于输入的样本,平衡类别之间的权重,下面写段测试代码测试这个函数:
# coding:utf-8 from sklearn.utils.class_weight import compute_class_weight class_weight = 'balanced' label = [0] * 9 + [1]*1 + [2, 2] print(label) # [0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 2, 2] classes=[0, 1, 2] weight = compute_class_weight(class_weight, classes, label) print(weight) #[ 0.44444444 4. 2. ] print(.44444444 * 9) # 3.99999996 print(4 * 1) # 4 print(2 * 2) # 4
如上图所示,可以看到这个函数把样本的平衡后的权重乘积为4,每个类别均如此。banlanced的计算公式为:n_samples/n_classes/np.bincount(y)。n_samples表示样本总数,n_classes表示总类别数量,np.bincount(y)输出所有类别的每个类别的样本数量,y是所有样本的标签。一个标签代表一个类别。采用balanced模型时,每种类别的权重为n_samples/n_classes,即12/3=4;然后根据每种类别中的样本数量对每个样本进行平均分配权重,即4/9=0.444, 4/1=4, 4/2=2。0类别有9个样本,1类别有1个样本,2类别有2个样本。
#calculate class weights
class_weights = class_weight.compute_class_weight( class_weight ='balanced',
classes =np.unique(y_train),
y =y_train.flatten())
Type: module
String form: <module 'sklearn.utils.class_weight' from '/home/software/anaconda3/envs/tf115/lib/python3.7/site-packages/sklearn/utils/class_weight.py'>
sklearn.utils.class_weight.compute_class_weight
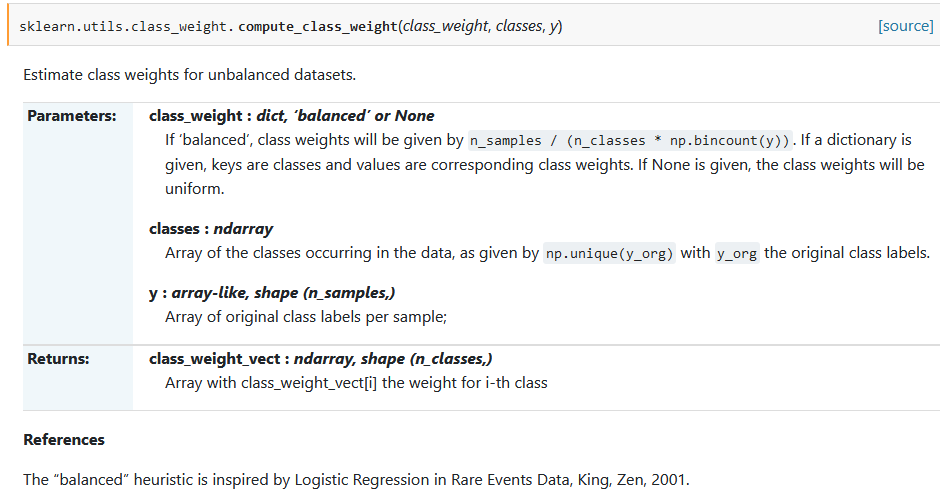
sklearn.utils.class_weight.compute_class_weight(class_weight, classes, y)[source]-
Estimate class weights for unbalanced datasets.
- Parameters
- class_weight dict, ‘balanced’ or None
-
If ‘balanced’, class weights will be given by
n_samples / (n_classes * np.bincount(y)). If a dictionary is given, keys are classes and values are corresponding class weights. If None is given, the class weights will be uniform. - classes ndarray
-
Array of the classes occurring in the data, as given by
np.unique(y_org)withy_orgthe original class labels. - y array-like, shape (n_samples,)
-
Array of original class labels per sample;
- Returns
- class_weight_vectndarray, shape (n_classes,)
-
Array with class_weight_vect[i] the weight for i-th class
References
The “balanced” heuristic is inspired by Logistic Regression in Rare Events Data, King, Zen, 2001.
REF
https://scikit-learn.org/0.22/modules/generated/sklearn.utils.class_weight.compute_class_weight.html
==================================================================================
import tensorflow as tf
import keras.backend as K
from keras.models import Sequential, Model
from keras.layers import Input, Dense, Dropout, Conv2D, MaxPooling2D, BatchNormalization, Flatten, GlobalAveragePooling2D, Multiply
from keras.callbacks import EarlyStopping, ModelCheckpoint
from keras.optimizers import Adam
from keras import backend as K
from keras.engine.topology import Layer, InputSpec
from keras.utils import Sequence, plot_model
from keras.constraints import unit_norm
from keras import regularizers
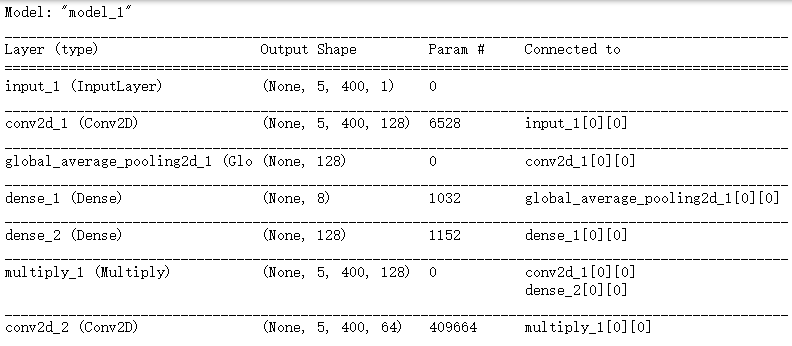
def SqueezeExcite(tensor, ratio=16):
nb_channel = K.int_shape(tensor)[-1] # 获得tensor的形状; -1 表示最后一个维度的长度,此处指图像的通道(5,400,1)
x = GlobalAveragePooling2D()(tensor) ## 每个通道进行求和平均;实际上只有一个通道;
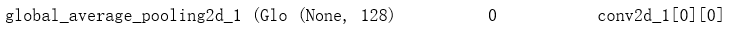
x = Dense(nb_channel // ratio, activation='relu')(x)

x = Dense(nb_channel, activation='sigmoid')(x)
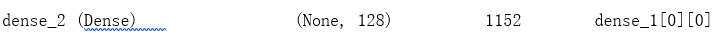
x = Multiply()([tensor, x])
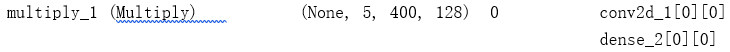
return x
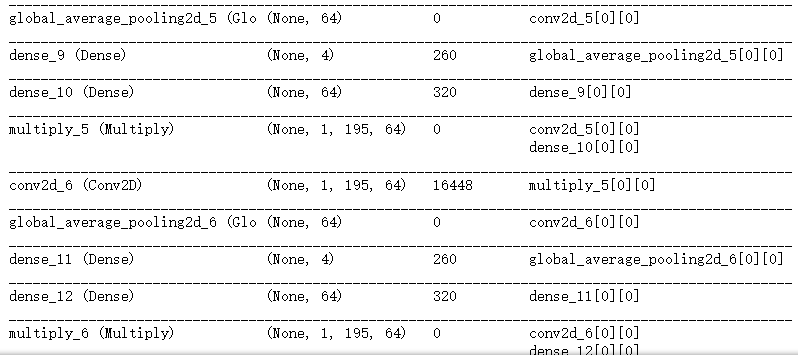
def create_model(width=200): ## width=400
K.clear_session() ## import keras.backend as K
pool2_list = []
merge_list = []
input_size = Input(shape=(5, width, 1)) ## 输入层 5,400,1 图像 宽 高 通道 keras.layers.Input
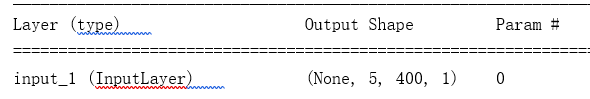
conv1_ = Conv2D(128, (5, 10), padding='same',activation='relu')(input_size) ## 128个核; 5*10大小的核;“same”代表保留边界处的卷积结果,通常会导致输出shape与输入shape相同。
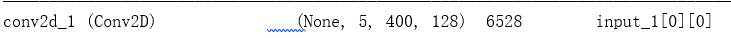
conv1 = SqueezeExcite(conv1_)
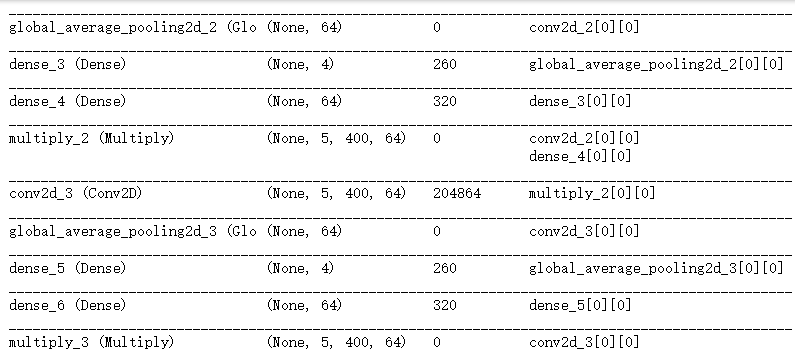
conv2_ = Conv2D(64, (5, 10), padding='same',activation='relu')(conv1)
conv2 = SqueezeExcite(conv2_)
conv3_ = Conv2D(64, (5, 10), padding='same',activation='relu')(conv2)
conv3 = SqueezeExcite(conv3_)
conv4_ = Conv2D(128, (5, 10), padding='valid',activation='relu')(conv3)
conv4 = SqueezeExcite(conv4_)
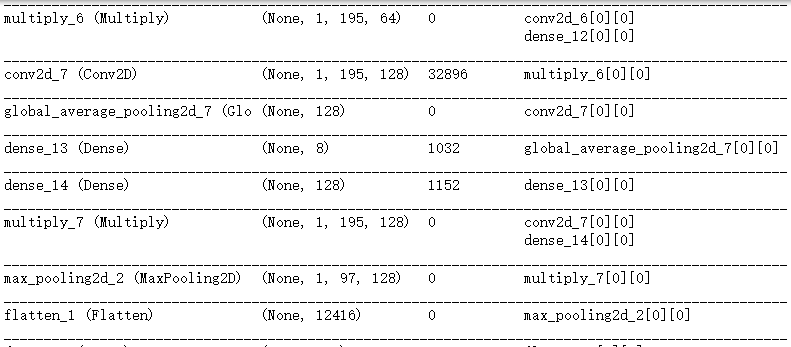
pool1 = MaxPooling2D(pool_size=(1, 2))(conv4)
conv5_ = Conv2D(64, (1, 4), padding='same',activation='relu')(pool1)
conv5 = SqueezeExcite(conv5_)
conv6_ = Conv2D(64, (1, 4), padding='same',activation='relu')(conv5)
conv6 = SqueezeExcite(conv6_)
conv7_ = Conv2D(128, (1, 4), padding='same',activation='relu')(conv6)
conv7 = SqueezeExcite(conv7_)
pool2 = MaxPooling2D(pool_size=(1, 2))(conv7)
x = Flatten()(pool2)
dense1 = Dense(256, activation='relu')(x)
x = Dropout(0.4)(dense1)
pred_output = Dense(1, activation='sigmoid')(x)
model = Model(input=[input_size], output=[pred_output])
model.summary()
return model
==================================================================================
Init signature:
EarlyStopping(
monitor='val_loss',
min_delta=0,
patience=0,
verbose=0,
mode='auto',
baseline=None,
restore_best_weights=False,
)
Docstring:
Stop training when a monitored quantity has stopped improving.
# Arguments
monitor: quantity to be monitored.
min_delta: minimum change in the monitored quantity
to qualify as an improvement, i.e. an absolute
change of less than min_delta, will count as no
improvement.
patience: number of epochs that produced the monitored
quantity with no improvement after which training will
be stopped.
Validation quantities may not be produced for every
epoch, if the validation frequency
(`model.fit(validation_freq=5)`) is greater than one.
verbose: verbosity mode.
mode: one of {auto, min, max}. In `min` mode,
training will stop when the quantity
monitored has stopped decreasing; in `max`
mode it will stop when the quantity
monitored has stopped increasing; in `auto`
mode, the direction is automatically inferred
from the name of the monitored quantity.
baseline: Baseline value for the monitored quantity to reach.
Training will stop if the model doesn't show improvement
over the baseline.
restore_best_weights: whether to restore model weights from
the epoch with the best value of the monitored quantity.
If False, the model weights obtained at the last step of
training are used.
File: //anaconda3/envs/tf115/lib/python3.7/site-packages/keras/callbacks/callbacks.py
Type: type
==================================================================================
Init signature:
Adam(
learning_rate=0.001,
beta_1=0.9,
beta_2=0.999,
amsgrad=False,
**kwargs,
)
Docstring:
Adam optimizer.
Default parameters follow those provided in the original paper.
# Arguments
learning_rate: float >= 0. Learning rate.
beta_1: float, 0 < beta < 1. Generally close to 1.
beta_2: float, 0 < beta < 1. Generally close to 1.
amsgrad: boolean. Whether to apply the AMSGrad variant of this
algorithm from the paper "On the Convergence of Adam and
Beyond".
# References
- [Adam - A Method for Stochastic Optimization](
https://arxiv.org/abs/1412.6980v8)
- [On the Convergence of Adam and Beyond](
https://openreview.net/forum?id=ryQu7f-RZ)
File: /home/software/anaconda3/envs/tf115/lib/python3.7/site-packages/keras/optimizers.py
Type: type
Subclasses:
==================================================================================
Signature: compile(source, filename, mode, flags=0, dont_inherit=False, optimize=-1)
Docstring:
Compile source into a code object that can be executed by exec() or eval().
The source code may represent a Python module, statement or expression.
The filename will be used for run-time error messages.
The mode must be 'exec' to compile a module, 'single' to compile a
single (interactive) statement, or 'eval' to compile an expression.
The flags argument, if present, controls which future statements influence
the compilation of the code.
The dont_inherit argument, if true, stops the compilation inheriting
the effects of any future statements in effect in the code calling
compile; if absent or false these statements do influence the compilation,
in addition to any features explicitly specified.
Type: builtin_function_or_method
==================================================================================
==================================================================================
==================================================================================
# construct the model
model = create_model(width=int(window_size/10))
es = EarlyStopping(monitor='val_loss', patience=2, restore_best_weights=True)
adam = Adam(lr=5e-5, beta_1=0.9, beta_2=0.999, epsilon=1e-08, decay=9e-5)
model.compile(loss='binary_crossentropy', optimizer=adam,
metrics=['accuracy', auroc, auprc, f1_m, recall_m, precision_m])
if os.path.exists('./saved_models/DNase_hg38.v8.h5'):
model.load_weights('./saved_models/DNase_hg38.v8.h5')
else:
#train the model
history = model.fit(x_train, y_train,
batch_size=32,
epochs=100,
validation_split=0.1,
shuffle=True,
class_weight=class_weights,
callbacks=[es])
model.save_weights('./saved_models/DNase_hg38.v8.h5')
==================================================================================


==================================================================================


 浙公网安备 33010602011771号
浙公网安备 33010602011771号