这个包依赖极有可能是这个:https://www.kegg.jp/kegg/docs/keggapi.html ,如果可以看懂会很好理解
由于KEGG数据库分享数据的策略改变,因此KEGG.db包不在能用,推荐KEGGREST包
But a number of years ago,KEGG changed their policy about sharing their data and so the KEGG.db package is no longer allowed to be current. Users who are interested in a more current pathway data are encouraged to look at the KEGGREST or reactome.db packages.
1、安装
if("KEGGREST" %in% rownames(installed.packages()) == FALSE) {source("http://bioconductor.org/biocLite.R");biocLite("KEGGREST")}
suppressMessages(library(KEGGREST))
ls('package:KEGGREST')
2、所有的对象及功能
keggConv:Convert KEGG identifiers to/from outside identifiers(ID转换功能)
keggFind:Finds entries with matching query keywords or other query data in a given database(即搜索功能)
keggGet:Retrieves given database entries(获取序列,图片等功能)
keggInfo:Displays the current statistics of a given database(即统计功能)
keggLink:Find related entries by using database cross-references(交互功能).
keggList:Returns a list of entry identifiers and associated definition for a given database or a given set of database entries.
listDatabases:Lists the KEGG databases which may be searched.
mark.pathway.by.objects:Client-side interface to obtain an url for a KEGG pathway diagram with a given set of genes marked(标记功能)
3、每个对象的功能简单使用
3.1、keggConv(Convert KEGG identifiers to/from outside identifiers,即ID转换功能)
语法:keggConv(target, source, querySize = 100)
例子:
## conversion from NCBI GeneID to KEGG ID for E. coli genes##
head(keggConv("eco", "ncbi-geneid"),2)
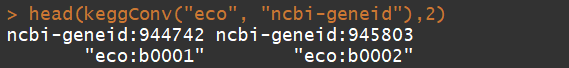
head(keggConv("ncbi-geneid", "eco"),2)

########conversion from KEGG ID to NCBI GI##########
head(keggConv("ncbi-proteinid", c("hsa:10458", "ece:Z5100")),2)

3.2)keggFind(Finds entries with matching query keywords or other query data in a given database,即检索功能)
语法:keggFind(database, query, option = c("formula", "exact_mass", "mol_weight"))
例子:
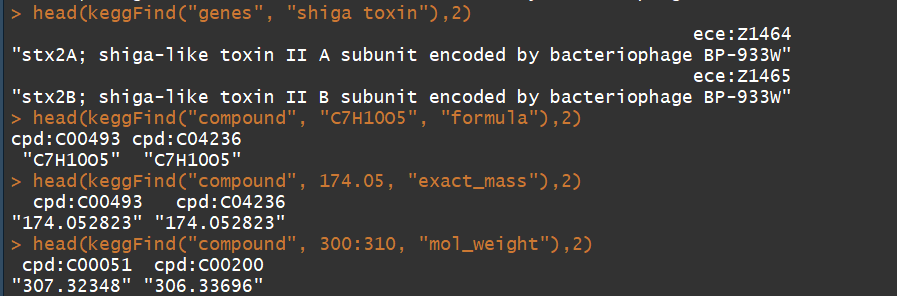
keggFind("genes", c("shiga", "toxin")) ## for keywords "shiga" and "toxin"
keggFind("genes", "shiga toxin") ## for keywords "shiga toxin"
keggFind("compound", "C7H10O5", "formula") ## for chemical formula "C7H10O5"
keggFind("compound", 174.05, "exact_mass") ## for 174.045 =< exact mass < 174.055
keggFind("compound", 300:310, "mol_weight") ## molecular weight 300 =< 310
3.3)keggGet (Retrieves given database entries,获取序列,图片等功能)
语法:keggGet(dbentries, option = c("aaseq", "ntseq", "mol", "kcf","image", "kgml"))
keggGet(c("cpd:C01290", "gl:G00092")) ## retrieves a compound entry and a glycan entry
keggGet(c("C01290", "G00092")) ## same as above, without prefixes
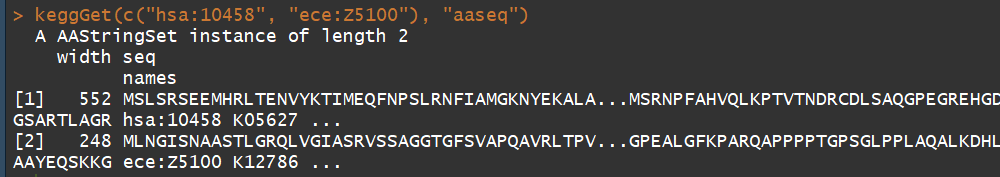
keggGet(c("hsa:10458", "ece:Z5100")) ## 检索 a human gene entry and an E.coli O157 gene entry
keggGet(c("hsa:10458", "ece:Z5100"), "aaseq") ## 检索 amino acid sequences of a human gene and an E.coli O157 gene
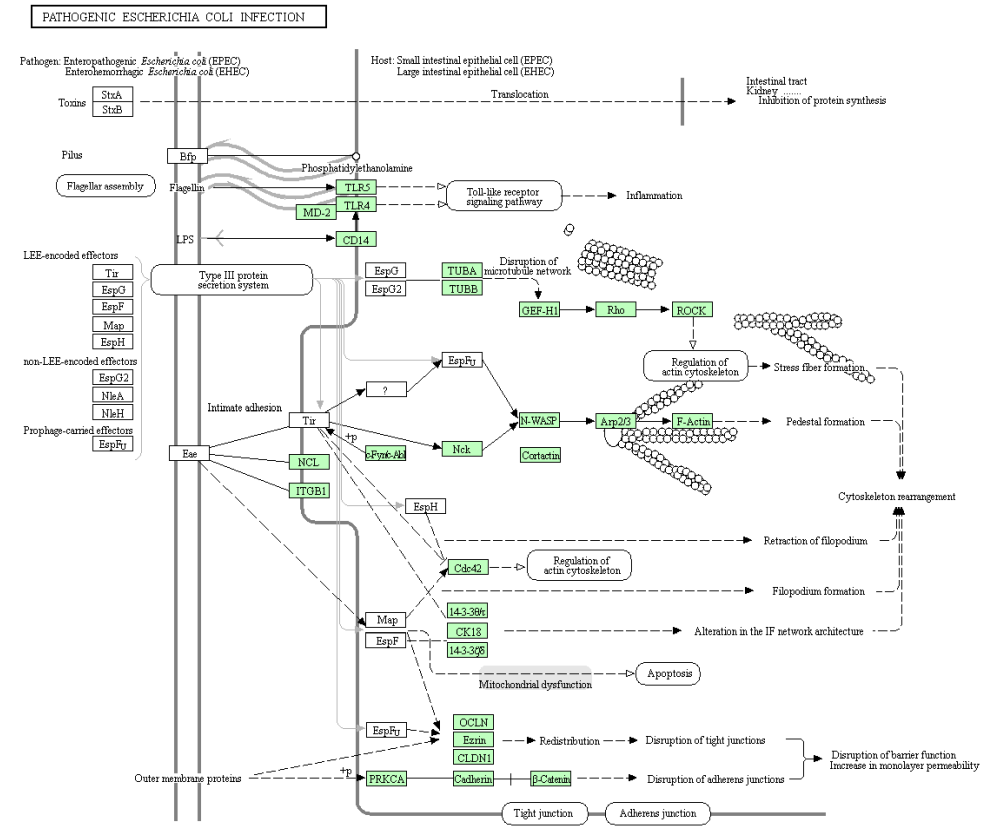
png <- keggGet("hsa05130", "image") ## retrieves the image file of a pathway map
t <- 'hsa05130.png'
library(png)
writePNG(png, t)
keggGet("hsa05130", "kgml")
hsa05130.png
3.4)keggInfo(Displays the current statistics of a given database,即统计功能)
语法:keggInfo(database)
head(keggInfo("kegg") ## displays the current statistics of the KEGG database
keggInfo("pathway") ## displays the number pathway entries including both the reference and organism-specific pathways
keggInfo("hsa") ##### displays the number of gene entries for the KEGG organism Homo sapiens
3.5)keggLink(keggLink Find related entries by using database cross-references.交互功能)
语法:keggLink(target, source)
head(keggLink("pathway", "hsa"),5) ### KEGG pathways linked from each of the human genes
head(keggLink("hsa", "pathway"),5) ## human genes linked from each of the KEGG pathways
head(keggLink("pathway", c("hsa:10458", "ece:Z5100")),5) ## KEGG pathways linked from a human gene and an E. coli O157 gene
head(keggLink("hsa:126"),5)

3.6) keggList (Returns a list of entry identifiers and associated definition for a given database or set of database entries,返回信息表)
用法:keggList(database, organism)
keggList("pathway") ## returns the list of reference pathways
keggList("pathway", "hsa") ## returns the list of human pathways
keggList("organism") ## returns the list of KEGG organisms with taxonomic classification
keggList("hsa") ## returns the entire list of human genes
keggList("T01001") ## same as above
keggList(c("hsa:10458", "ece:Z5100")) ## returns the list of a human gene and an E.coli O157 gene
keggList(c("cpd:C01290","gl:G00092")) ## returns the list of a compound entry and a glycan entry
keggList(c("C01290+G00092")) ## same as above (prefixes are not necessary)
3.7) listDatabases(Lists the KEGG databases which may be searched.查看有数据库)
用法:listDatabases()
例子:
listDatabases() ##查看有哪些数据库
3.8)mark.pathway.by.objects:Client-side interface to obtain an url for a KEGG pathway diagram with a given set of genes marked(标记功能,例如上调y用红框,下调用绿框)
语法:
mark.pathway.by.objects(pathway.id, object.id.list)
color.pathway.by.objects(pathway.id, object.id.list,fg.color.list, bg.color.list)
例子:
url <- mark.pathway.by.objects("path:eco00260",c("eco:b0002", "eco:c00263"))
if(interactive()){browseURL(url)}
url <- color.pathway.by.objects("path:eco00260",c("eco:b0002", "eco:c00263"),c("#ff0000", "#00ff00"), c("#ffff00", "yellow"))



 浙公网安备 33010602011771号
浙公网安备 33010602011771号