相似的基因在不同物种中,其功能往往保守的。显然,需要一个统一的术语用于描述这些跨物种的同源基因及其基因产物的功能,否则,不同的实验室对相同的基因的功能的描述不同,将极大限制学术的交流。而 Gene Ontology (GO) 项目正是为了能够使对各种数据库中基因获基因产物功能描述相一致的努力结果。一般情况下,在各种组学中,做完差异表达之后,会看这些差异表达基因参与的功能,一般我们会做一个GO注释和富集。这里我们先来了解一下GO.db这个包。因为该包是许多其他包的基础依赖包。对于了解GO有很大的帮助。
1)安装载入
-------------------------------------------
if("GO.db" %in% rownames(installed.packages()) == FALSE) {source("http://bioconductor.org/biocLite.R");biocLite("GO.db")}
suppressMessages(library(GO.db))
2)查看该包所有的对象
--------------------------------------------
ls("package:GO.db")
![]()
columns(GO.db)
"DEFINITION" "GOID" "ONTOLOGY" "TERM"
功能:可以用来进行GO的注释
GOBPANCESTOR:Annotation of GO Identifiers to their Biological Process Ancestors
GOBPCHILDREN:Annotation of GO Identifiers to their Biological Process Children
GOBPOFFSPRING: Annotation of GO Identifiers to their Biological Process Offspring
GOBPPARENTS: Annotation of GO Identifiers to their Biological Process Parents
GOCCANCESTOR: Annotation of GO Identifiers to their Cellular Component Ancestors
GOCCCHILDREN: Annotation of GO Identifiers to their Cellular Component Children
GOCCOFFSPRING: Annotation of GO Identifiers to their Cellular Component Offspring
GOCCPARENTS: Annotation of GO Identifiers to their Cellular Component Parents
GOMAPCOUNTS Number of mapped keys for the maps in package GO.db
GOMFANCESTOR: Annotation of GO identifiers to their Molecular Function Ancestors
GOMFCHILDREN: Annotation of GO Identifiers to their Molecular Function Children
GOMFOFFSPRING: Annotation of GO Identifiers to their Molecular Function Offspring
GOMFPARENTS: Annotation of GO Identifiers to their Molecular Function Parents
GOOBSOLETE: Annotation of GO identifiers by terms defined by Gene Ontology Consortium and their status are obsolete
GOSYNONYM:Map from GO synonyms to GO terms
GOTERM: Annotation of GO Identifiers to GO Terms
GO_dbconn:Collect information about the package: Annotation DB
原理:所依赖的数据库ftp://ftp.geneontology.org/pub/go/godatabase/archive/latest-lite/
3)各个对象的简单使用
-----------------------------------------------------------
3.1)GOBPANCESTOR (Annotation of GO Identifiers to all of their Biological Process Ancestors,即返回的是该GO号所有的长辈关系)
xx <- as.list(GOBPANCESTOR) # Convert the object to a list
xx <- xx[!is.na(xx)] # Remove GO IDs that do not have any ancestor
if(length(xx) > 0){
goids <- xx[1] # Get the ancestor GO IDs for the first two elents of xx
}
goids
$`GO:0000001`
[1] "GO:0006996" "GO:0007005" "GO:0008150" "GO:0009987" "GO:0016043" "GO:0048308"
[7] "GO:0048311" "GO:0051179" "GO:0051640" "GO:0051641" "GO:0051646" "GO:0071840"
[13] "all"
xx <- as.list(GOBPCHILDREN)
xx <- xx[!is.na(xx)] # Remove GO IDs that do not have any children
if(length(xx) > 0){
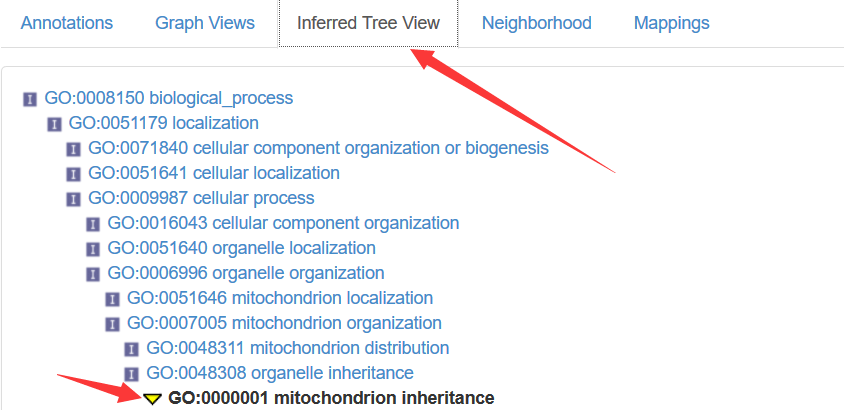
goids <- xx[[1]] # Get the parent GO IDs for the first elents of xx part_of "GO:0032042" ; is_a "GO:0033955"
GOID(GOTERM[[goids[1]]]) #"GO:0032042"
Term(GOTERM[[goids[1]]]) #"mitochondrial DNA metabolic process"
Synonym(GOTERM[[goids[1]]]) #"mitochondrial DNA metabolism" "mtDNA metabolic process" "mtDNA metabolism"
Secondary(GOTERM[[goids[1]]]) #character(0)
Definition(GOTERM[[goids[1]]]) # "The chemical reactions and pathways involving mitochondrial DNA."
Ontology(GOTERM[[goids[1]]]) #"BP"
}
3.3)GOBPOFFSPRING(Annotation of GO Identifiers to their Biological Process Offspring,子孙后裔,即相当于所有有血缘关系)
xx <- as.list(GOBPOFFSPRING)
xx <- xx[!is.na(xx)] # Remove GO IDs that do not have any offspring
if(length(xx) > 0){
goids <- xx[1] # Get the offspring GO IDs for the first of xx
}
goids
$`GO:0000002`
[1] "GO:0006264" "GO:0032042" "GO:0032043" "GO:0033955" "GO:0043504" "GO:0090296"
[7] "GO:0090297" "GO:0090298" "GO:1901858" "GO:1901859" "GO:1901860" "GO:1905951"

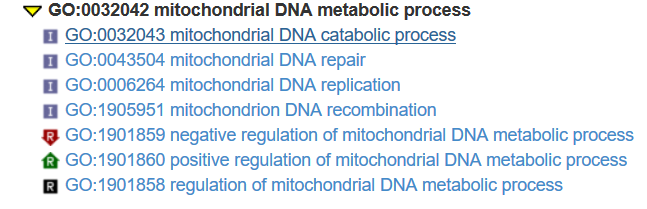
![]()
3.4) GOBPPARENTS(Annotation of GO Identifiers to their Biological Process direct Parents,即返回的是直接父亲)
xx <- as.list(GOBPPARENTS)
xx <- xx[!is.na(xx)] # Remove GO IDs that do not have any parent
if(length(xx) > 0){
goids <- xx[[1]] # Get the children GO IDs for the first elents of xx #is_a "GO:0048308" ,is_a "GO:0048311"
GOID(GOTERM[[goids[1]]]) # Find out the GO terms for the first parent goid #"GO:0048308"
Term(GOTERM[[goids[1]]]) #"organelle inheritance"
Synonym(GOTERM[[goids[1]]]) #character(0)
Secondary(GOTERM[[goids[1]]]) #character(0)
Definition(GOTERM[[goids[1]]]) #"The partitioning of organelles between daughter cells at cell division."
Ontology(GOTERM[[goids[1]]]) #"BP"
}
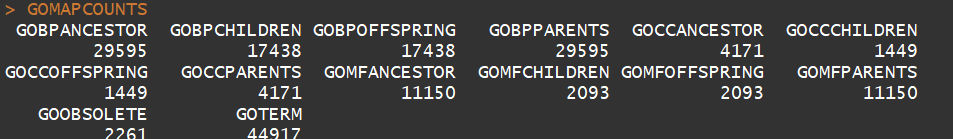
3.5)GOMAPCOUNTS (Number of mapped keys for the maps in package GO.db)
GOMAPCOUNTS
3.6)GOOBSOLETE(Annotation of GO identifiers by terms defined by Gene Ontology Consortium and their status are obsolete)
xx <- as.list(GOTERM)
if(length(xx) > 0){
GOID(xx[[1]]) # Get the TERMS for the first elent of xx "GO:0000001"
Ontology(xx[[1]]) "BP"
}
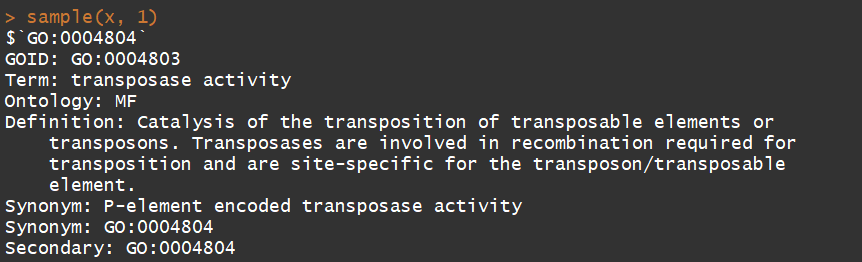
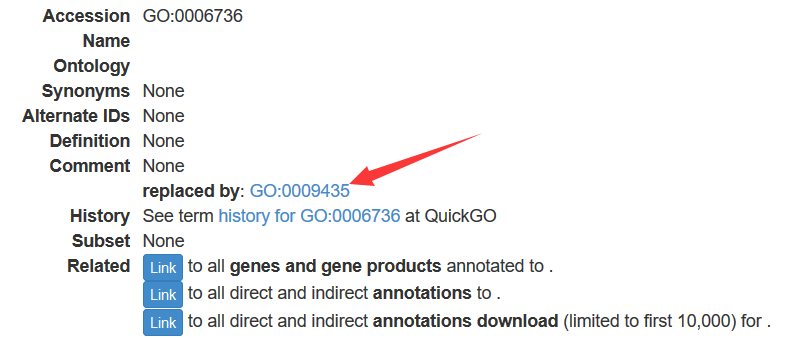
x <- GOSYNONYM sample(x, 1) GOTERM[["GO:0009435"]] # GO ID "GO:0009435" has a lot of synonyms GOID(GOSYNONYM[["GO:0006736"]]) # GO ID "GO:0006736" is a synonym of GO ID "GO:0009435"
3.8)GOTERM Annotation of GO Identifiers to GO Terms
xx <- as.list(GOTERM)
if(length(xx) > 0){
GOID(xx[[1]]) ##"GO:0000001"
Term(xx[[1]]) ##"mitochondrion inheritance"
Synonym(xx[[1]]) #"mitochondrial inheritance"
Secondary(xx[[1]]) ##character(0)
Definition(xx[[1]]) #"The distribution of mitochondria, including the mitochondrial genome, into daughter cells after mitosis or meiosis, mediated by interactions between mitochondria and the cytoskeleton.
Ontology(xx[[1]]) #"BP"
}
3.9)GO_dbconn(Collect information about the package annotation DB)
GO_dbconn()
GO_dbfile()
GO_dbInfo()
这里以BP为例,CC,MF和其它道理相同